
Deinococcus irradiatisoli
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Deinococcus-Thermus; Deinococci; Deinococcales; Deinococcaceae; Deinococcus
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
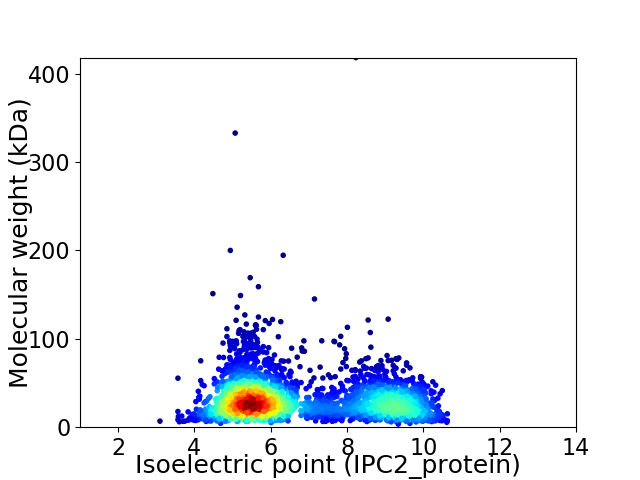
Virtual 2D-PAGE plot for 2903 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z3JFY9|A0A2Z3JFY9_9DEIO Carboxylic ester hydrolase OS=Deinococcus irradiatisoli OX=2202254 GN=DKM44_13425 PE=3 SV=1
MM1 pKa = 6.96NTPSEE6 pKa = 4.03LRR8 pKa = 11.84YY9 pKa = 10.14APSHH13 pKa = 5.94EE14 pKa = 4.17WLSDD18 pKa = 3.35DD19 pKa = 3.67GTVGISDD26 pKa = 4.46FAQDD30 pKa = 3.29QLGDD34 pKa = 3.68VVYY37 pKa = 11.21VEE39 pKa = 4.37LPEE42 pKa = 4.24VGRR45 pKa = 11.84EE46 pKa = 4.01VQAGEE51 pKa = 4.07TVAVVEE57 pKa = 4.56SVKK60 pKa = 10.0TASDD64 pKa = 3.45IYY66 pKa = 11.02APASGTITAVNEE78 pKa = 3.96ALAGSPEE85 pKa = 4.46LVNGDD90 pKa = 4.1CYY92 pKa = 11.57GEE94 pKa = 4.15GWLFKK99 pKa = 10.85LDD101 pKa = 3.56VTAEE105 pKa = 4.1SGDD108 pKa = 3.99LLDD111 pKa = 4.11AASYY115 pKa = 9.39EE116 pKa = 4.35AANSS120 pKa = 3.58
MM1 pKa = 6.96NTPSEE6 pKa = 4.03LRR8 pKa = 11.84YY9 pKa = 10.14APSHH13 pKa = 5.94EE14 pKa = 4.17WLSDD18 pKa = 3.35DD19 pKa = 3.67GTVGISDD26 pKa = 4.46FAQDD30 pKa = 3.29QLGDD34 pKa = 3.68VVYY37 pKa = 11.21VEE39 pKa = 4.37LPEE42 pKa = 4.24VGRR45 pKa = 11.84EE46 pKa = 4.01VQAGEE51 pKa = 4.07TVAVVEE57 pKa = 4.56SVKK60 pKa = 10.0TASDD64 pKa = 3.45IYY66 pKa = 11.02APASGTITAVNEE78 pKa = 3.96ALAGSPEE85 pKa = 4.46LVNGDD90 pKa = 4.1CYY92 pKa = 11.57GEE94 pKa = 4.15GWLFKK99 pKa = 10.85LDD101 pKa = 3.56VTAEE105 pKa = 4.1SGDD108 pKa = 3.99LLDD111 pKa = 4.11AASYY115 pKa = 9.39EE116 pKa = 4.35AANSS120 pKa = 3.58
Molecular weight: 12.59 kDa
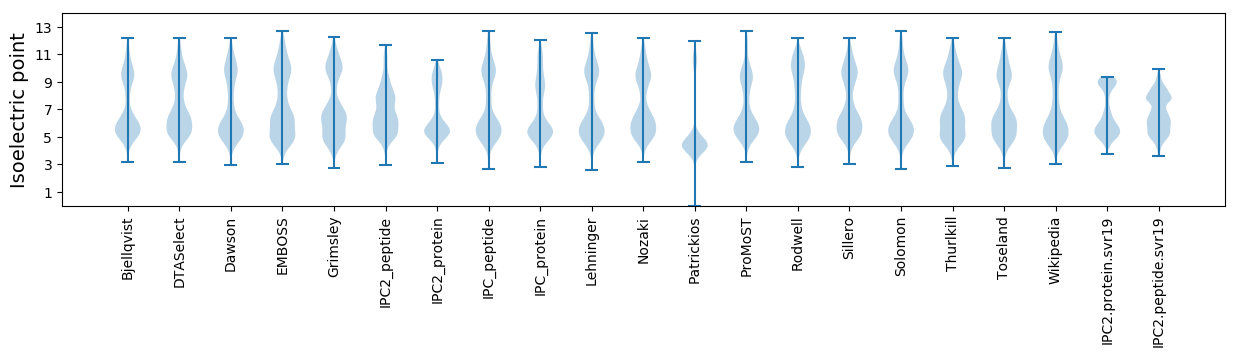
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z3JEN9|A0A2Z3JEN9_9DEIO Uncharacterized protein OS=Deinococcus irradiatisoli OX=2202254 GN=DKM44_09505 PE=4 SV=1
MM1 pKa = 7.51SAPHH5 pKa = 6.7LADD8 pKa = 3.14ATLRR12 pKa = 11.84AYY14 pKa = 10.48ALGDD18 pKa = 3.76LPLPAQQAAEE28 pKa = 4.15GHH30 pKa = 5.82VLGCPLCRR38 pKa = 11.84AQVIRR43 pKa = 11.84WRR45 pKa = 11.84DD46 pKa = 3.3DD47 pKa = 3.04FAAQVDD53 pKa = 4.2ALPSAGRR60 pKa = 11.84LPPLRR65 pKa = 11.84RR66 pKa = 11.84PHH68 pKa = 6.21GAPAKK73 pKa = 9.95RR74 pKa = 11.84HH75 pKa = 5.43LPPRR79 pKa = 11.84WSALTLPGLLMLSVLALGWGGWQQHH104 pKa = 4.94QASALRR110 pKa = 11.84AEE112 pKa = 4.39QRR114 pKa = 11.84QVAGWLAQPQLKK126 pKa = 9.38VLAMQDD132 pKa = 3.25TKK134 pKa = 10.92RR135 pKa = 11.84RR136 pKa = 11.84PTGQLLLLPSRR147 pKa = 11.84EE148 pKa = 4.09VLFVLPPPAPGKK160 pKa = 10.52VYY162 pKa = 9.6QVWVAAHH169 pKa = 5.47WHH171 pKa = 6.47RR172 pKa = 11.84GDD174 pKa = 3.73KK175 pKa = 9.21LTPQLSSRR183 pKa = 11.84RR184 pKa = 11.84GTFSASVGEE193 pKa = 3.91NDD195 pKa = 3.9YY196 pKa = 11.56VCVSLEE202 pKa = 3.97DD203 pKa = 4.18AGRR206 pKa = 11.84QPTKK210 pKa = 9.95PSRR213 pKa = 11.84ILGWSMVV220 pKa = 3.33
MM1 pKa = 7.51SAPHH5 pKa = 6.7LADD8 pKa = 3.14ATLRR12 pKa = 11.84AYY14 pKa = 10.48ALGDD18 pKa = 3.76LPLPAQQAAEE28 pKa = 4.15GHH30 pKa = 5.82VLGCPLCRR38 pKa = 11.84AQVIRR43 pKa = 11.84WRR45 pKa = 11.84DD46 pKa = 3.3DD47 pKa = 3.04FAAQVDD53 pKa = 4.2ALPSAGRR60 pKa = 11.84LPPLRR65 pKa = 11.84RR66 pKa = 11.84PHH68 pKa = 6.21GAPAKK73 pKa = 9.95RR74 pKa = 11.84HH75 pKa = 5.43LPPRR79 pKa = 11.84WSALTLPGLLMLSVLALGWGGWQQHH104 pKa = 4.94QASALRR110 pKa = 11.84AEE112 pKa = 4.39QRR114 pKa = 11.84QVAGWLAQPQLKK126 pKa = 9.38VLAMQDD132 pKa = 3.25TKK134 pKa = 10.92RR135 pKa = 11.84RR136 pKa = 11.84PTGQLLLLPSRR147 pKa = 11.84EE148 pKa = 4.09VLFVLPPPAPGKK160 pKa = 10.52VYY162 pKa = 9.6QVWVAAHH169 pKa = 5.47WHH171 pKa = 6.47RR172 pKa = 11.84GDD174 pKa = 3.73KK175 pKa = 9.21LTPQLSSRR183 pKa = 11.84RR184 pKa = 11.84GTFSASVGEE193 pKa = 3.91NDD195 pKa = 3.9YY196 pKa = 11.56VCVSLEE202 pKa = 3.97DD203 pKa = 4.18AGRR206 pKa = 11.84QPTKK210 pKa = 9.95PSRR213 pKa = 11.84ILGWSMVV220 pKa = 3.33
Molecular weight: 24.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
896936 |
33 |
4079 |
309.0 |
33.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.258 ± 0.065 | 0.555 ± 0.013 |
4.887 ± 0.033 | 5.633 ± 0.05 |
3.189 ± 0.027 | 8.982 ± 0.041 |
2.082 ± 0.023 | 3.61 ± 0.038 |
2.849 ± 0.044 | 12.047 ± 0.074 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.84 ± 0.021 | 2.516 ± 0.031 |
5.697 ± 0.042 | 4.271 ± 0.039 |
6.941 ± 0.052 | 5.694 ± 0.039 |
5.495 ± 0.037 | 7.625 ± 0.031 |
1.389 ± 0.022 | 2.439 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
