
Ruminococcus albus SY3
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Ruminococcus; Ruminococcus albus
Average proteome isoelectric point is 5.81
Get precalculated fractions of proteins
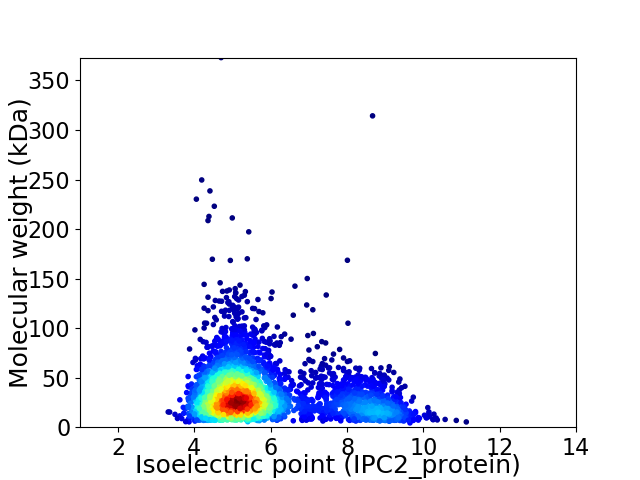
Virtual 2D-PAGE plot for 3322 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
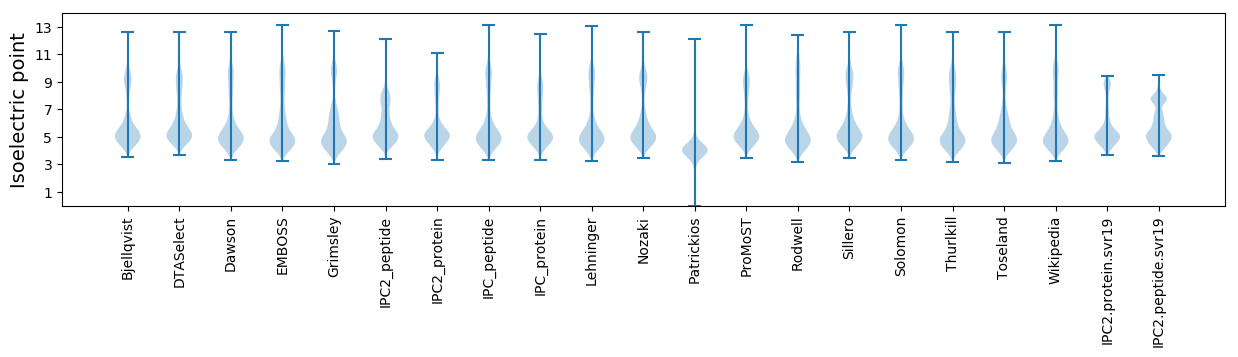
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A011VVA6|A0A011VVA6_RUMAL Diguanylate cyclase OS=Ruminococcus albus SY3 OX=1341156 GN=RASY3_04210 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 10.13PIIKK6 pKa = 10.03RR7 pKa = 11.84LLSGVLSSTMTISAIPIVSAHH28 pKa = 6.57ADD30 pKa = 3.62EE31 pKa = 5.01SAEE34 pKa = 4.16SYY36 pKa = 10.07PYY38 pKa = 11.37TMFAASNDD46 pKa = 3.71DD47 pKa = 3.57GAITINADD55 pKa = 3.38NFCVNGNVATNGTIVSSGNMNINGTKK81 pKa = 9.61TEE83 pKa = 4.19SADD86 pKa = 3.14EE87 pKa = 4.08SMIFIFDD94 pKa = 4.58KK95 pKa = 10.77IDD97 pKa = 3.1TKK99 pKa = 11.24YY100 pKa = 10.75FSVPNVEE107 pKa = 4.45EE108 pKa = 3.89HH109 pKa = 6.21TEE111 pKa = 4.08DD112 pKa = 3.61YY113 pKa = 10.95TIDD116 pKa = 3.63EE117 pKa = 4.69LNININVPTEE127 pKa = 4.14VQGEE131 pKa = 4.14ATLTGNININNALKK145 pKa = 10.82ALDD148 pKa = 4.02DD149 pKa = 3.96VNLYY153 pKa = 11.32GEE155 pKa = 4.55VKK157 pKa = 9.23NTNDD161 pKa = 3.57SIIFSKK167 pKa = 11.17YY168 pKa = 8.81GDD170 pKa = 3.39IVIDD174 pKa = 3.8SQNVNLNGLIYY185 pKa = 10.62APFGSVTINAQNLNLNNVVIIAEE208 pKa = 4.62SIVLTCPNVNANNGSNASSFVGTSSEE234 pKa = 4.18TFIAPIEE241 pKa = 3.93EE242 pKa = 3.95WQYY245 pKa = 11.85LNDD248 pKa = 3.78NDD250 pKa = 4.35EE251 pKa = 5.11DD252 pKa = 4.78GIPDD256 pKa = 3.72LVEE259 pKa = 3.86NAIGTDD265 pKa = 3.41NDD267 pKa = 4.04SQDD270 pKa = 3.4SDD272 pKa = 3.64GDD274 pKa = 4.06NIPDD278 pKa = 3.37YY279 pKa = 11.1TEE281 pKa = 4.24YY282 pKa = 11.29VVLNTDD288 pKa = 3.19PTSPNDD294 pKa = 3.37VMPFIQAAYY303 pKa = 10.03DD304 pKa = 3.52NLEE307 pKa = 3.99IEE309 pKa = 4.48LAIGDD314 pKa = 4.05TYY316 pKa = 11.97DD317 pKa = 3.3NVNNDD322 pKa = 2.47VVLPLKK328 pKa = 9.67STYY331 pKa = 10.51DD332 pKa = 3.4AVVEE336 pKa = 4.22WEE338 pKa = 4.08VSNDD342 pKa = 2.91EE343 pKa = 4.31VININGHH350 pKa = 5.57ISDD353 pKa = 4.16NLEE356 pKa = 3.95SSSEE360 pKa = 3.96VTYY363 pKa = 9.48TATIFLFGEE372 pKa = 4.07QLTKK376 pKa = 10.85KK377 pKa = 10.33FDD379 pKa = 3.38LHH381 pKa = 6.56VNPKK385 pKa = 10.07ASHH388 pKa = 6.62IDD390 pKa = 2.96IDD392 pKa = 4.41SIKK395 pKa = 10.87DD396 pKa = 3.46LTVDD400 pKa = 5.31DD401 pKa = 4.41IEE403 pKa = 4.83EE404 pKa = 4.19MNSDD408 pKa = 4.08DD409 pKa = 4.31EE410 pKa = 4.76DD411 pKa = 4.61FEE413 pKa = 5.86IEE415 pKa = 4.11INDD418 pKa = 3.36YY419 pKa = 11.26GYY421 pKa = 10.27IEE423 pKa = 5.08EE424 pKa = 4.68IFGKK428 pKa = 10.91YY429 pKa = 9.65SDD431 pKa = 4.61NSIKK435 pKa = 10.85SPNDD439 pKa = 3.27ALYY442 pKa = 10.9SLYY445 pKa = 10.63SIKK448 pKa = 9.79TALGISDD455 pKa = 4.06PFEE458 pKa = 3.89EE459 pKa = 5.25LKK461 pKa = 11.23LFDD464 pKa = 4.05IDD466 pKa = 3.93KK467 pKa = 10.96DD468 pKa = 3.87EE469 pKa = 4.52TGTIYY474 pKa = 11.1KK475 pKa = 7.99FTQLVNGVPCFDD487 pKa = 3.85NNIIVSCDD495 pKa = 3.2EE496 pKa = 4.49NGTTDD501 pKa = 3.77YY502 pKa = 11.07LRR504 pKa = 11.84SSYY507 pKa = 10.67FPVNTTISTSPNISYY522 pKa = 11.02EE523 pKa = 4.34DD524 pKa = 3.56INDD527 pKa = 5.23DD528 pKa = 3.93ILKK531 pKa = 9.33EE532 pKa = 4.05YY533 pKa = 9.76PDD535 pKa = 3.86ASIIDD540 pKa = 3.53VDD542 pKa = 3.51EE543 pKa = 4.81DD544 pKa = 3.38NRR546 pKa = 11.84LFILNYY552 pKa = 9.22YY553 pKa = 9.45GKK555 pKa = 10.9LYY557 pKa = 10.58LVWNKK562 pKa = 10.6YY563 pKa = 10.05IGVNNYY569 pKa = 8.05TYY571 pKa = 10.7QILVNANDD579 pKa = 3.88GKK581 pKa = 10.61ILYY584 pKa = 10.3KK585 pKa = 10.65NISDD589 pKa = 3.78INFDD593 pKa = 3.72EE594 pKa = 5.43DD595 pKa = 3.96LTITQDD601 pKa = 4.06DD602 pKa = 4.32LLKK605 pKa = 10.34QSRR608 pKa = 11.84TISILKK614 pKa = 9.53QNNLFVDD621 pKa = 4.17DD622 pKa = 4.53YY623 pKa = 11.52YY624 pKa = 11.84YY625 pKa = 11.36LEE627 pKa = 5.11DD628 pKa = 3.47SDD630 pKa = 5.72RR631 pKa = 11.84NIIVYY636 pKa = 9.49DD637 pKa = 3.97AQGKK641 pKa = 10.35AYY643 pKa = 10.22GSSDD647 pKa = 2.74KK648 pKa = 11.36KK649 pKa = 10.82NIKK652 pKa = 9.52KK653 pKa = 9.9HH654 pKa = 5.65LFLFIIILVHH664 pKa = 6.67HH665 pKa = 6.87GLLKK669 pKa = 10.49KK670 pKa = 10.64YY671 pKa = 10.18LLWQTWKK678 pKa = 10.07IYY680 pKa = 9.86MIFTITNSVDD690 pKa = 3.9FLLMMLGLKK699 pKa = 7.52QQEE702 pKa = 4.54MII704 pKa = 4.28
MM1 pKa = 7.35KK2 pKa = 10.13PIIKK6 pKa = 10.03RR7 pKa = 11.84LLSGVLSSTMTISAIPIVSAHH28 pKa = 6.57ADD30 pKa = 3.62EE31 pKa = 5.01SAEE34 pKa = 4.16SYY36 pKa = 10.07PYY38 pKa = 11.37TMFAASNDD46 pKa = 3.71DD47 pKa = 3.57GAITINADD55 pKa = 3.38NFCVNGNVATNGTIVSSGNMNINGTKK81 pKa = 9.61TEE83 pKa = 4.19SADD86 pKa = 3.14EE87 pKa = 4.08SMIFIFDD94 pKa = 4.58KK95 pKa = 10.77IDD97 pKa = 3.1TKK99 pKa = 11.24YY100 pKa = 10.75FSVPNVEE107 pKa = 4.45EE108 pKa = 3.89HH109 pKa = 6.21TEE111 pKa = 4.08DD112 pKa = 3.61YY113 pKa = 10.95TIDD116 pKa = 3.63EE117 pKa = 4.69LNININVPTEE127 pKa = 4.14VQGEE131 pKa = 4.14ATLTGNININNALKK145 pKa = 10.82ALDD148 pKa = 4.02DD149 pKa = 3.96VNLYY153 pKa = 11.32GEE155 pKa = 4.55VKK157 pKa = 9.23NTNDD161 pKa = 3.57SIIFSKK167 pKa = 11.17YY168 pKa = 8.81GDD170 pKa = 3.39IVIDD174 pKa = 3.8SQNVNLNGLIYY185 pKa = 10.62APFGSVTINAQNLNLNNVVIIAEE208 pKa = 4.62SIVLTCPNVNANNGSNASSFVGTSSEE234 pKa = 4.18TFIAPIEE241 pKa = 3.93EE242 pKa = 3.95WQYY245 pKa = 11.85LNDD248 pKa = 3.78NDD250 pKa = 4.35EE251 pKa = 5.11DD252 pKa = 4.78GIPDD256 pKa = 3.72LVEE259 pKa = 3.86NAIGTDD265 pKa = 3.41NDD267 pKa = 4.04SQDD270 pKa = 3.4SDD272 pKa = 3.64GDD274 pKa = 4.06NIPDD278 pKa = 3.37YY279 pKa = 11.1TEE281 pKa = 4.24YY282 pKa = 11.29VVLNTDD288 pKa = 3.19PTSPNDD294 pKa = 3.37VMPFIQAAYY303 pKa = 10.03DD304 pKa = 3.52NLEE307 pKa = 3.99IEE309 pKa = 4.48LAIGDD314 pKa = 4.05TYY316 pKa = 11.97DD317 pKa = 3.3NVNNDD322 pKa = 2.47VVLPLKK328 pKa = 9.67STYY331 pKa = 10.51DD332 pKa = 3.4AVVEE336 pKa = 4.22WEE338 pKa = 4.08VSNDD342 pKa = 2.91EE343 pKa = 4.31VININGHH350 pKa = 5.57ISDD353 pKa = 4.16NLEE356 pKa = 3.95SSSEE360 pKa = 3.96VTYY363 pKa = 9.48TATIFLFGEE372 pKa = 4.07QLTKK376 pKa = 10.85KK377 pKa = 10.33FDD379 pKa = 3.38LHH381 pKa = 6.56VNPKK385 pKa = 10.07ASHH388 pKa = 6.62IDD390 pKa = 2.96IDD392 pKa = 4.41SIKK395 pKa = 10.87DD396 pKa = 3.46LTVDD400 pKa = 5.31DD401 pKa = 4.41IEE403 pKa = 4.83EE404 pKa = 4.19MNSDD408 pKa = 4.08DD409 pKa = 4.31EE410 pKa = 4.76DD411 pKa = 4.61FEE413 pKa = 5.86IEE415 pKa = 4.11INDD418 pKa = 3.36YY419 pKa = 11.26GYY421 pKa = 10.27IEE423 pKa = 5.08EE424 pKa = 4.68IFGKK428 pKa = 10.91YY429 pKa = 9.65SDD431 pKa = 4.61NSIKK435 pKa = 10.85SPNDD439 pKa = 3.27ALYY442 pKa = 10.9SLYY445 pKa = 10.63SIKK448 pKa = 9.79TALGISDD455 pKa = 4.06PFEE458 pKa = 3.89EE459 pKa = 5.25LKK461 pKa = 11.23LFDD464 pKa = 4.05IDD466 pKa = 3.93KK467 pKa = 10.96DD468 pKa = 3.87EE469 pKa = 4.52TGTIYY474 pKa = 11.1KK475 pKa = 7.99FTQLVNGVPCFDD487 pKa = 3.85NNIIVSCDD495 pKa = 3.2EE496 pKa = 4.49NGTTDD501 pKa = 3.77YY502 pKa = 11.07LRR504 pKa = 11.84SSYY507 pKa = 10.67FPVNTTISTSPNISYY522 pKa = 11.02EE523 pKa = 4.34DD524 pKa = 3.56INDD527 pKa = 5.23DD528 pKa = 3.93ILKK531 pKa = 9.33EE532 pKa = 4.05YY533 pKa = 9.76PDD535 pKa = 3.86ASIIDD540 pKa = 3.53VDD542 pKa = 3.51EE543 pKa = 4.81DD544 pKa = 3.38NRR546 pKa = 11.84LFILNYY552 pKa = 9.22YY553 pKa = 9.45GKK555 pKa = 10.9LYY557 pKa = 10.58LVWNKK562 pKa = 10.6YY563 pKa = 10.05IGVNNYY569 pKa = 8.05TYY571 pKa = 10.7QILVNANDD579 pKa = 3.88GKK581 pKa = 10.61ILYY584 pKa = 10.3KK585 pKa = 10.65NISDD589 pKa = 3.78INFDD593 pKa = 3.72EE594 pKa = 5.43DD595 pKa = 3.96LTITQDD601 pKa = 4.06DD602 pKa = 4.32LLKK605 pKa = 10.34QSRR608 pKa = 11.84TISILKK614 pKa = 9.53QNNLFVDD621 pKa = 4.17DD622 pKa = 4.53YY623 pKa = 11.52YY624 pKa = 11.84YY625 pKa = 11.36LEE627 pKa = 5.11DD628 pKa = 3.47SDD630 pKa = 5.72RR631 pKa = 11.84NIIVYY636 pKa = 9.49DD637 pKa = 3.97AQGKK641 pKa = 10.35AYY643 pKa = 10.22GSSDD647 pKa = 2.74KK648 pKa = 11.36KK649 pKa = 10.82NIKK652 pKa = 9.52KK653 pKa = 9.9HH654 pKa = 5.65LFLFIIILVHH664 pKa = 6.67HH665 pKa = 6.87GLLKK669 pKa = 10.49KK670 pKa = 10.64YY671 pKa = 10.18LLWQTWKK678 pKa = 10.07IYY680 pKa = 9.86MIFTITNSVDD690 pKa = 3.9FLLMMLGLKK699 pKa = 7.52QQEE702 pKa = 4.54MII704 pKa = 4.28
Molecular weight: 78.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A011UF00|A0A011UF00_RUMAL 50S ribosomal protein L16 OS=Ruminococcus albus SY3 OX=1341156 GN=rplP PE=3 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.65QPKK8 pKa = 9.08KK9 pKa = 8.13LQRR12 pKa = 11.84KK13 pKa = 8.12KK14 pKa = 8.7VHH16 pKa = 5.82GFRR19 pKa = 11.84KK20 pKa = 10.07RR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.93VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84ARR41 pKa = 11.84LTHH44 pKa = 6.31
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.65QPKK8 pKa = 9.08KK9 pKa = 8.13LQRR12 pKa = 11.84KK13 pKa = 8.12KK14 pKa = 8.7VHH16 pKa = 5.82GFRR19 pKa = 11.84KK20 pKa = 10.07RR21 pKa = 11.84MATANGRR28 pKa = 11.84KK29 pKa = 8.93VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84ARR41 pKa = 11.84LTHH44 pKa = 6.31
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1072678 |
37 |
3489 |
322.9 |
36.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.562 ± 0.044 | 1.693 ± 0.02 |
6.697 ± 0.032 | 7.222 ± 0.052 |
4.245 ± 0.028 | 7.04 ± 0.045 |
1.6 ± 0.019 | 7.26 ± 0.038 |
7.037 ± 0.038 | 8.089 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.897 ± 0.025 | 4.749 ± 0.037 |
3.201 ± 0.025 | 2.451 ± 0.022 |
4.173 ± 0.038 | 6.421 ± 0.049 |
5.822 ± 0.054 | 6.664 ± 0.035 |
0.892 ± 0.016 | 4.286 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
