
Geobacillus phage GBK1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
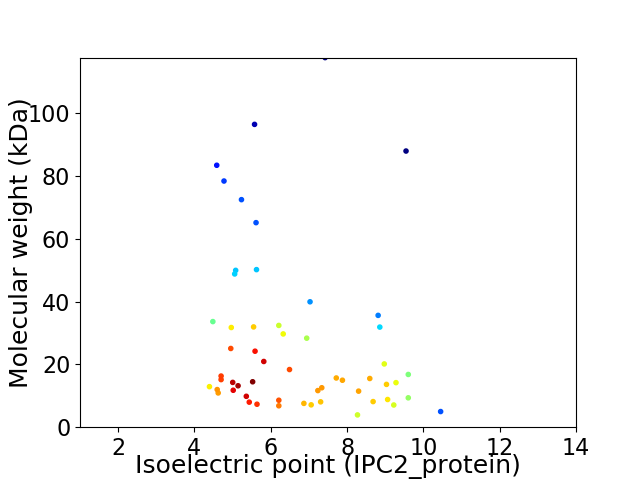
Virtual 2D-PAGE plot for 56 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
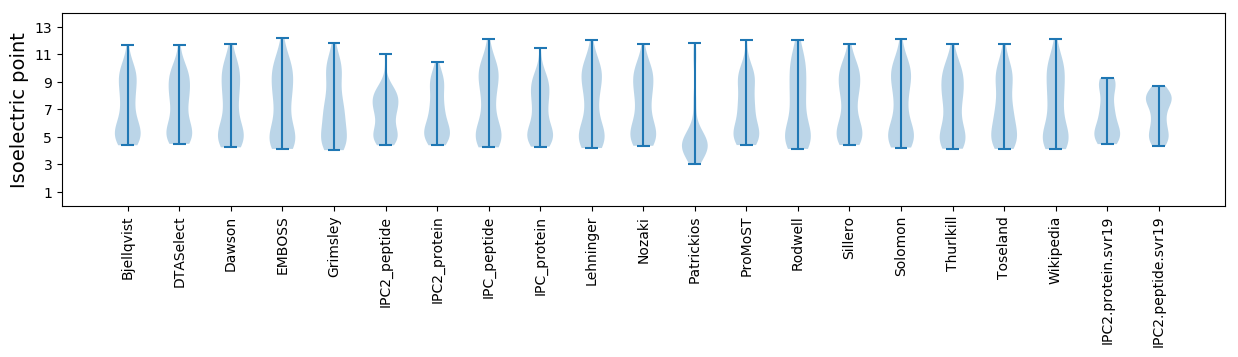
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6B9LX04|A0A6B9LX04_9CAUD DNA primase OS=Geobacillus phage GBK1 OX=2686286 GN=GBK1_26 PE=4 SV=1
MM1 pKa = 7.7PWYY4 pKa = 10.04KK5 pKa = 10.47YY6 pKa = 10.32LRR8 pKa = 11.84AYY10 pKa = 10.03QGLGTGWISFDD21 pKa = 3.35LTNVFMNDD29 pKa = 3.62YY30 pKa = 11.26KK31 pKa = 11.07NGTYY35 pKa = 10.12HH36 pKa = 7.2GVVLYY41 pKa = 10.8SGASSGYY48 pKa = 7.97YY49 pKa = 10.22GEE51 pKa = 4.99AYY53 pKa = 10.21GYY55 pKa = 8.39RR56 pKa = 11.84TDD58 pKa = 3.81SYY60 pKa = 10.48CAYY63 pKa = 10.9VEE65 pKa = 4.51VTGSWNSPPSAPTITYY81 pKa = 7.83PQGGEE86 pKa = 3.97IVDD89 pKa = 3.48QSITVRR95 pKa = 11.84WNKK98 pKa = 10.35ASDD101 pKa = 3.9PDD103 pKa = 4.26GDD105 pKa = 3.92PLSYY109 pKa = 10.48EE110 pKa = 4.09VAINDD115 pKa = 3.68GTGWKK120 pKa = 9.84YY121 pKa = 11.34YY122 pKa = 9.09KK123 pKa = 9.6TATGATSYY131 pKa = 10.29TINTSSLRR139 pKa = 11.84EE140 pKa = 3.78TSSARR145 pKa = 11.84VAVRR149 pKa = 11.84AYY151 pKa = 10.37DD152 pKa = 3.68GQEE155 pKa = 3.95YY156 pKa = 10.97SSWTYY161 pKa = 11.56SNYY164 pKa = 9.97FAIDD168 pKa = 3.41HH169 pKa = 6.1NQPPSAPTQLSPATGAVIDD188 pKa = 3.71RR189 pKa = 11.84TQPNRR194 pKa = 11.84FSWKK198 pKa = 10.23HH199 pKa = 5.57NDD201 pKa = 3.65DD202 pKa = 3.68GAQVGFRR209 pKa = 11.84IAWRR213 pKa = 11.84TVAADD218 pKa = 3.68GTRR221 pKa = 11.84GAWNYY226 pKa = 10.0IPNATSFMNTTNQYY240 pKa = 10.85YY241 pKa = 10.9DD242 pKa = 3.59MPANTLPFGNIEE254 pKa = 4.02WGVQTKK260 pKa = 9.88DD261 pKa = 3.35QQGLQSPYY269 pKa = 10.38SQWQIIKK276 pKa = 10.48ASQPTNAPTILSPTDD291 pKa = 3.47FAEE294 pKa = 4.22INTTRR299 pKa = 11.84VTVTWSSLNQIQYY312 pKa = 11.36DD313 pKa = 3.83LALYY317 pKa = 10.62DD318 pKa = 3.54SDD320 pKa = 3.21GWEE323 pKa = 4.26LFHH326 pKa = 6.55EE327 pKa = 4.91VKK329 pKa = 10.78SGGTKK334 pKa = 9.7QVTIPVDD341 pKa = 3.54LEE343 pKa = 4.02NNKK346 pKa = 9.83FYY348 pKa = 10.53EE349 pKa = 3.63IHH351 pKa = 6.39LRR353 pKa = 11.84VMDD356 pKa = 4.0STTGLWSDD364 pKa = 3.57YY365 pKa = 9.31TVVVFSTNFTPPLPPVIEE383 pKa = 4.28RR384 pKa = 11.84FEE386 pKa = 3.98EE387 pKa = 4.67AGDD390 pKa = 3.7GVINIFYY397 pKa = 9.9GASDD401 pKa = 4.06ADD403 pKa = 3.62ILPSFLVNDD412 pKa = 4.78GQQNPLLQGYY422 pKa = 8.58SGSVIGTDD430 pKa = 4.26CEE432 pKa = 4.25LLSPSSVSITGALKK446 pKa = 10.55GVEE449 pKa = 3.88YY450 pKa = 10.17WLTNEE455 pKa = 4.96EE456 pKa = 4.16IPLVAGSVYY465 pKa = 10.58TLTATFDD472 pKa = 3.65VQGGRR477 pKa = 11.84LFIGAYY483 pKa = 9.5DD484 pKa = 3.87ANNNAISFQATGADD498 pKa = 3.53KK499 pKa = 10.97ALSPVGTTALNYY511 pKa = 7.34TLPEE515 pKa = 4.01NAVKK519 pKa = 10.54LRR521 pKa = 11.84VIFYY525 pKa = 6.97TTWDD529 pKa = 3.33NTGTVTISNANLRR542 pKa = 11.84ISNASTATEE551 pKa = 4.5KK552 pKa = 10.48IQVFRR557 pKa = 11.84RR558 pKa = 11.84EE559 pKa = 3.98YY560 pKa = 9.83TPTGTAPWIMIADD573 pKa = 4.63DD574 pKa = 4.83LPTSGSFLDD583 pKa = 3.73YY584 pKa = 10.41TPASGVIYY592 pKa = 9.52EE593 pKa = 4.29YY594 pKa = 11.29KK595 pKa = 10.49LRR597 pKa = 11.84AVNNSNKK604 pKa = 9.44TSTDD608 pKa = 2.88SSIEE612 pKa = 3.74QVSIVFNDD620 pKa = 3.57TFIQEE625 pKa = 4.31ANNLSSIVLLKK636 pKa = 10.71YY637 pKa = 8.89ATSRR641 pKa = 11.84EE642 pKa = 4.05TKK644 pKa = 9.5MEE646 pKa = 4.25LEE648 pKa = 4.2SEE650 pKa = 4.34LMRR653 pKa = 11.84FAGRR657 pKa = 11.84RR658 pKa = 11.84DD659 pKa = 3.36PVRR662 pKa = 11.84EE663 pKa = 3.98FGEE666 pKa = 4.37HH667 pKa = 5.63EE668 pKa = 4.99DD669 pKa = 3.52IVISVEE675 pKa = 4.22WIFDD679 pKa = 3.57TYY681 pKa = 11.36QEE683 pKa = 4.14VEE685 pKa = 4.17MFKK688 pKa = 11.05DD689 pKa = 3.39LLRR692 pKa = 11.84RR693 pKa = 11.84RR694 pKa = 11.84DD695 pKa = 3.42VLLYY699 pKa = 9.92RR700 pKa = 11.84DD701 pKa = 3.5RR702 pKa = 11.84NGRR705 pKa = 11.84RR706 pKa = 11.84FWVTCDD712 pKa = 3.02AFEE715 pKa = 5.15VKK717 pKa = 10.75DD718 pKa = 3.61NDD720 pKa = 3.85VNGFTLKK727 pKa = 10.76ADD729 pKa = 3.41FHH731 pKa = 5.67VTHH734 pKa = 7.34FIEE737 pKa = 5.41DD738 pKa = 3.64LAVRR742 pKa = 11.84NGEE745 pKa = 4.13EE746 pKa = 3.88TT747 pKa = 3.31
MM1 pKa = 7.7PWYY4 pKa = 10.04KK5 pKa = 10.47YY6 pKa = 10.32LRR8 pKa = 11.84AYY10 pKa = 10.03QGLGTGWISFDD21 pKa = 3.35LTNVFMNDD29 pKa = 3.62YY30 pKa = 11.26KK31 pKa = 11.07NGTYY35 pKa = 10.12HH36 pKa = 7.2GVVLYY41 pKa = 10.8SGASSGYY48 pKa = 7.97YY49 pKa = 10.22GEE51 pKa = 4.99AYY53 pKa = 10.21GYY55 pKa = 8.39RR56 pKa = 11.84TDD58 pKa = 3.81SYY60 pKa = 10.48CAYY63 pKa = 10.9VEE65 pKa = 4.51VTGSWNSPPSAPTITYY81 pKa = 7.83PQGGEE86 pKa = 3.97IVDD89 pKa = 3.48QSITVRR95 pKa = 11.84WNKK98 pKa = 10.35ASDD101 pKa = 3.9PDD103 pKa = 4.26GDD105 pKa = 3.92PLSYY109 pKa = 10.48EE110 pKa = 4.09VAINDD115 pKa = 3.68GTGWKK120 pKa = 9.84YY121 pKa = 11.34YY122 pKa = 9.09KK123 pKa = 9.6TATGATSYY131 pKa = 10.29TINTSSLRR139 pKa = 11.84EE140 pKa = 3.78TSSARR145 pKa = 11.84VAVRR149 pKa = 11.84AYY151 pKa = 10.37DD152 pKa = 3.68GQEE155 pKa = 3.95YY156 pKa = 10.97SSWTYY161 pKa = 11.56SNYY164 pKa = 9.97FAIDD168 pKa = 3.41HH169 pKa = 6.1NQPPSAPTQLSPATGAVIDD188 pKa = 3.71RR189 pKa = 11.84TQPNRR194 pKa = 11.84FSWKK198 pKa = 10.23HH199 pKa = 5.57NDD201 pKa = 3.65DD202 pKa = 3.68GAQVGFRR209 pKa = 11.84IAWRR213 pKa = 11.84TVAADD218 pKa = 3.68GTRR221 pKa = 11.84GAWNYY226 pKa = 10.0IPNATSFMNTTNQYY240 pKa = 10.85YY241 pKa = 10.9DD242 pKa = 3.59MPANTLPFGNIEE254 pKa = 4.02WGVQTKK260 pKa = 9.88DD261 pKa = 3.35QQGLQSPYY269 pKa = 10.38SQWQIIKK276 pKa = 10.48ASQPTNAPTILSPTDD291 pKa = 3.47FAEE294 pKa = 4.22INTTRR299 pKa = 11.84VTVTWSSLNQIQYY312 pKa = 11.36DD313 pKa = 3.83LALYY317 pKa = 10.62DD318 pKa = 3.54SDD320 pKa = 3.21GWEE323 pKa = 4.26LFHH326 pKa = 6.55EE327 pKa = 4.91VKK329 pKa = 10.78SGGTKK334 pKa = 9.7QVTIPVDD341 pKa = 3.54LEE343 pKa = 4.02NNKK346 pKa = 9.83FYY348 pKa = 10.53EE349 pKa = 3.63IHH351 pKa = 6.39LRR353 pKa = 11.84VMDD356 pKa = 4.0STTGLWSDD364 pKa = 3.57YY365 pKa = 9.31TVVVFSTNFTPPLPPVIEE383 pKa = 4.28RR384 pKa = 11.84FEE386 pKa = 3.98EE387 pKa = 4.67AGDD390 pKa = 3.7GVINIFYY397 pKa = 9.9GASDD401 pKa = 4.06ADD403 pKa = 3.62ILPSFLVNDD412 pKa = 4.78GQQNPLLQGYY422 pKa = 8.58SGSVIGTDD430 pKa = 4.26CEE432 pKa = 4.25LLSPSSVSITGALKK446 pKa = 10.55GVEE449 pKa = 3.88YY450 pKa = 10.17WLTNEE455 pKa = 4.96EE456 pKa = 4.16IPLVAGSVYY465 pKa = 10.58TLTATFDD472 pKa = 3.65VQGGRR477 pKa = 11.84LFIGAYY483 pKa = 9.5DD484 pKa = 3.87ANNNAISFQATGADD498 pKa = 3.53KK499 pKa = 10.97ALSPVGTTALNYY511 pKa = 7.34TLPEE515 pKa = 4.01NAVKK519 pKa = 10.54LRR521 pKa = 11.84VIFYY525 pKa = 6.97TTWDD529 pKa = 3.33NTGTVTISNANLRR542 pKa = 11.84ISNASTATEE551 pKa = 4.5KK552 pKa = 10.48IQVFRR557 pKa = 11.84RR558 pKa = 11.84EE559 pKa = 3.98YY560 pKa = 9.83TPTGTAPWIMIADD573 pKa = 4.63DD574 pKa = 4.83LPTSGSFLDD583 pKa = 3.73YY584 pKa = 10.41TPASGVIYY592 pKa = 9.52EE593 pKa = 4.29YY594 pKa = 11.29KK595 pKa = 10.49LRR597 pKa = 11.84AVNNSNKK604 pKa = 9.44TSTDD608 pKa = 2.88SSIEE612 pKa = 3.74QVSIVFNDD620 pKa = 3.57TFIQEE625 pKa = 4.31ANNLSSIVLLKK636 pKa = 10.71YY637 pKa = 8.89ATSRR641 pKa = 11.84EE642 pKa = 4.05TKK644 pKa = 9.5MEE646 pKa = 4.25LEE648 pKa = 4.2SEE650 pKa = 4.34LMRR653 pKa = 11.84FAGRR657 pKa = 11.84RR658 pKa = 11.84DD659 pKa = 3.36PVRR662 pKa = 11.84EE663 pKa = 3.98FGEE666 pKa = 4.37HH667 pKa = 5.63EE668 pKa = 4.99DD669 pKa = 3.52IVISVEE675 pKa = 4.22WIFDD679 pKa = 3.57TYY681 pKa = 11.36QEE683 pKa = 4.14VEE685 pKa = 4.17MFKK688 pKa = 11.05DD689 pKa = 3.39LLRR692 pKa = 11.84RR693 pKa = 11.84RR694 pKa = 11.84DD695 pKa = 3.42VLLYY699 pKa = 9.92RR700 pKa = 11.84DD701 pKa = 3.5RR702 pKa = 11.84NGRR705 pKa = 11.84RR706 pKa = 11.84FWVTCDD712 pKa = 3.02AFEE715 pKa = 5.15VKK717 pKa = 10.75DD718 pKa = 3.61NDD720 pKa = 3.85VNGFTLKK727 pKa = 10.76ADD729 pKa = 3.41FHH731 pKa = 5.67VTHH734 pKa = 7.34FIEE737 pKa = 5.41DD738 pKa = 3.64LAVRR742 pKa = 11.84NGEE745 pKa = 4.13EE746 pKa = 3.88TT747 pKa = 3.31
Molecular weight: 83.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6B9LPB7|A0A6B9LPB7_9CAUD Uncharacterized protein OS=Geobacillus phage GBK1 OX=2686286 GN=GBK1_10 PE=4 SV=1
MM1 pKa = 7.43NKK3 pKa = 9.66LHH5 pKa = 6.53LHH7 pKa = 5.71VDD9 pKa = 3.41STKK12 pKa = 10.7KK13 pKa = 9.85KK14 pKa = 10.34RR15 pKa = 11.84KK16 pKa = 7.8GQLWCPYY23 pKa = 8.84CGEE26 pKa = 3.7WTYY29 pKa = 11.55FKK31 pKa = 10.25MFRR34 pKa = 11.84KK35 pKa = 9.73LRR37 pKa = 11.84FFPFQSTYY45 pKa = 10.07RR46 pKa = 11.84RR47 pKa = 11.84CVKK50 pKa = 10.47CGISVEE56 pKa = 4.26DD57 pKa = 3.88FWVKK61 pKa = 9.06TVNKK65 pKa = 9.34LWSFGVNSRR74 pKa = 11.84RR75 pKa = 11.84KK76 pKa = 9.24
MM1 pKa = 7.43NKK3 pKa = 9.66LHH5 pKa = 6.53LHH7 pKa = 5.71VDD9 pKa = 3.41STKK12 pKa = 10.7KK13 pKa = 9.85KK14 pKa = 10.34RR15 pKa = 11.84KK16 pKa = 7.8GQLWCPYY23 pKa = 8.84CGEE26 pKa = 3.7WTYY29 pKa = 11.55FKK31 pKa = 10.25MFRR34 pKa = 11.84KK35 pKa = 9.73LRR37 pKa = 11.84FFPFQSTYY45 pKa = 10.07RR46 pKa = 11.84RR47 pKa = 11.84CVKK50 pKa = 10.47CGISVEE56 pKa = 4.26DD57 pKa = 3.88FWVKK61 pKa = 9.06TVNKK65 pKa = 9.34LWSFGVNSRR74 pKa = 11.84RR75 pKa = 11.84KK76 pKa = 9.24
Molecular weight: 9.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13209 |
33 |
1028 |
235.9 |
26.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.7 ± 0.491 | 0.931 ± 0.164 |
5.496 ± 0.292 | 8.577 ± 0.589 |
4.467 ± 0.231 | 6.405 ± 0.259 |
1.832 ± 0.204 | 6.337 ± 0.228 |
7.987 ± 0.555 | 8.714 ± 0.325 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.392 ± 0.146 | 4.671 ± 0.23 |
3.407 ± 0.179 | 3.838 ± 0.21 |
5.443 ± 0.27 | 5.193 ± 0.533 |
5.368 ± 0.427 | 6.98 ± 0.323 |
1.219 ± 0.125 | 4.043 ± 0.261 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
