
uncultured bacterium
Taxonomy: cellular organisms; Bacteria; environmental samples
Average proteome isoelectric point is 7.08
Get precalculated fractions of proteins
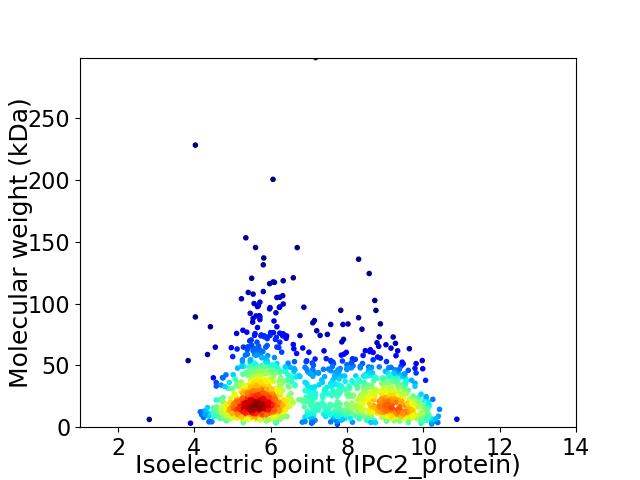
Virtual 2D-PAGE plot for 1290 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
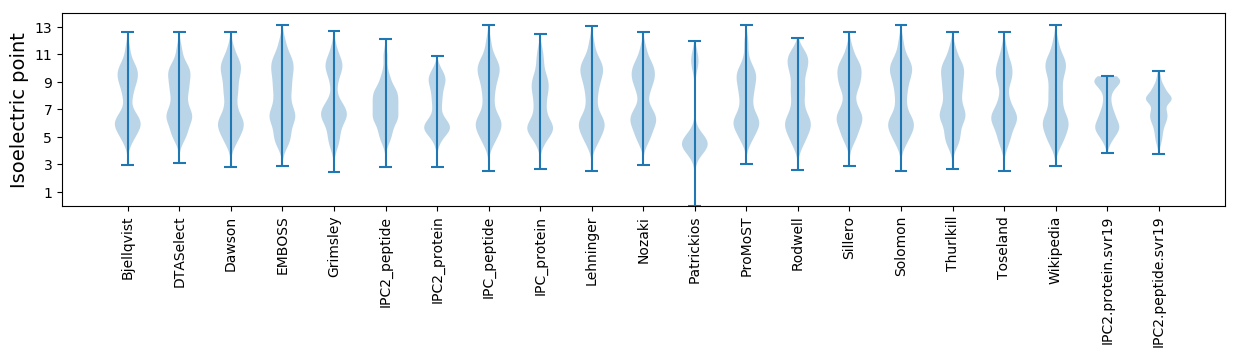
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A090KNA6|A0A090KNA6_9BACT Uncultured bacterium genome assembly Metasoil_fosmids_resub OS=uncultured bacterium OX=77133 PE=4 SV=1
MM1 pKa = 7.13IRR3 pKa = 11.84APRR6 pKa = 11.84TRR8 pKa = 11.84RR9 pKa = 11.84SFNRR13 pKa = 11.84SRR15 pKa = 11.84LWLEE19 pKa = 3.74SLEE22 pKa = 4.27TRR24 pKa = 11.84VTPTTIPVSTLNDD37 pKa = 3.34MGQGSLRR44 pKa = 11.84QAIIDD49 pKa = 3.75ANFNPGPDD57 pKa = 3.47TIEE60 pKa = 3.83FTVAGTIQLTTGALPTITDD79 pKa = 3.61TVKK82 pKa = 10.21IDD84 pKa = 3.23GRR86 pKa = 11.84SAPGFLAAPVVEE98 pKa = 4.49IDD100 pKa = 3.09AHH102 pKa = 5.87GFAGLFFEE110 pKa = 5.09IGSNGSSLLSLAVGNANGAGVTLDD134 pKa = 3.84DD135 pKa = 4.91SNNNVAGNYY144 pKa = 9.32IGLALDD150 pKa = 4.55GSTTASNVGDD160 pKa = 3.42GLVINSSSINNIIGGTSPLDD180 pKa = 3.43RR181 pKa = 11.84NVISGNTGDD190 pKa = 4.86GITVSGSTRR199 pKa = 11.84NQIVGNFIGPDD210 pKa = 3.2ASGLGGRR217 pKa = 11.84GNGLDD222 pKa = 4.91GILLTAGAAVNNVGGTIDD240 pKa = 3.39AQRR243 pKa = 11.84NIISCNAADD252 pKa = 4.56GVSLEE257 pKa = 4.45QGAHH261 pKa = 6.43DD262 pKa = 5.68NIILGNFIGTDD273 pKa = 3.44VTGLAGLGNGLDD285 pKa = 4.0GVHH288 pKa = 7.23ADD290 pKa = 3.75DD291 pKa = 5.29AANNVIGNTVPVKK304 pKa = 10.46NVSYY308 pKa = 10.61FNANLVSPTPNPGWTGIRR326 pKa = 11.84ASDD329 pKa = 3.26TSGNYY334 pKa = 9.86LISGISGTQGLLYY347 pKa = 10.48EE348 pKa = 4.58GTISGAGVTIPVNFPNASASAVYY371 pKa = 10.47GPDD374 pKa = 3.22NLGGNHH380 pKa = 5.27VALVGSYY387 pKa = 10.36NVTGTGLADD396 pKa = 3.13QKK398 pKa = 11.65GFIFEE403 pKa = 4.2GTTADD408 pKa = 3.48LTNPANYY415 pKa = 7.33TTIDD419 pKa = 3.04AGGDD423 pKa = 3.49FNIVHH428 pKa = 6.08STMQGLAVGNSDD440 pKa = 3.63SSPSQGQASLLSHH453 pKa = 6.95AFLYY457 pKa = 10.7DD458 pKa = 3.42VASGQFLPDD467 pKa = 2.86IVFPGSVSNTAYY479 pKa = 10.49GIWFNGGTSYY489 pKa = 9.53TICGGWSSDD498 pKa = 3.41PVNNFDD504 pKa = 4.94DD505 pKa = 3.76QNRR508 pKa = 11.84PIGQGYY514 pKa = 8.82LVDD517 pKa = 4.35YY518 pKa = 11.02DD519 pKa = 4.8SLTQTFTNWTSFQYY533 pKa = 10.73PFGKK537 pKa = 10.28SFFTHH542 pKa = 6.54FEE544 pKa = 4.55GISSVEE550 pKa = 3.87NGVYY554 pKa = 9.5TLNADD559 pKa = 3.72SFQVGTTNPIQGSWVSVRR577 pKa = 11.84RR578 pKa = 11.84NSDD581 pKa = 2.78GSFSLGQWTNLAYY594 pKa = 9.88PGTGNITSSNSVYY607 pKa = 10.13GNQVVGVVFHH617 pKa = 6.86GATAFAFQATVNIGFQLSNVIGCNADD643 pKa = 3.7DD644 pKa = 4.7GVLLTGGATANTIASNFIGLDD665 pKa = 3.49LTGKK669 pKa = 10.14AALANGSDD677 pKa = 3.63GVEE680 pKa = 4.21VEE682 pKa = 4.11NSQGNLIGTTSTGPEE697 pKa = 3.39VNYY700 pKa = 10.31FNTSLITSVPVSGWEE715 pKa = 4.56GIRR718 pKa = 11.84QADD721 pKa = 3.98LPGQFLISGTSGSNGLLFEE740 pKa = 4.19GTIDD744 pKa = 4.62GVGTTNIVDD753 pKa = 3.64YY754 pKa = 9.98PGASSTFVYY763 pKa = 10.62GPDD766 pKa = 3.57NEE768 pKa = 4.73GSGNLGLVGTYY779 pKa = 7.92KK780 pKa = 10.07TANSSVVNGFLFQGTVADD798 pKa = 5.16LGTPANFQTINKK810 pKa = 9.71SGATFNYY817 pKa = 9.54LHH819 pKa = 6.28STMGGLIVGNYY830 pKa = 9.93DD831 pKa = 3.15SAPDD835 pKa = 3.72HH836 pKa = 6.18GQGGLPFGPGHH847 pKa = 5.64ATIYY851 pKa = 10.71DD852 pKa = 3.69IATQTYY858 pKa = 7.89LTDD861 pKa = 3.19IVFPGSKK868 pKa = 9.98SNSVYY873 pKa = 10.87GIWFNGGTSYY883 pKa = 9.4TVCGGYY889 pKa = 10.12SPTFVNNFDD898 pKa = 3.81DD899 pKa = 3.72QNRR902 pKa = 11.84PIGQAYY908 pKa = 9.02LANYY912 pKa = 7.69DD913 pKa = 3.65TATGQFSHH921 pKa = 6.68FTSFSYY927 pKa = 10.7PFGTNFLTHH936 pKa = 6.55FEE938 pKa = 4.92GISSVEE944 pKa = 3.78PGVYY948 pKa = 9.24TLAADD953 pKa = 4.9SIQSGSSQTIQASWVVVRR971 pKa = 11.84RR972 pKa = 11.84NPDD975 pKa = 2.68GSFGTGQWVNLDD987 pKa = 3.67YY988 pKa = 11.2PGVATTGTFTGSDD1001 pKa = 3.25SVYY1004 pKa = 9.18GNQVVGIVVSSTSTFCFQATVNVGTQMSNIISGNSGNGVLLNVGAQNNQVAGNFIGTDD1062 pKa = 3.36ATGSASLGNSLNGVQLEE1079 pKa = 4.37GANNNVIGNNNPITDD1094 pKa = 3.37IPFFNTDD1101 pKa = 3.04SVSVQVSGWQGIRR1114 pKa = 11.84NSDD1117 pKa = 3.3TTGQFLITGTSGTNGLLFDD1136 pKa = 3.87GTIAGVGTSYY1146 pKa = 10.87QVNFPNAATTSVYY1159 pKa = 10.81GPDD1162 pKa = 3.48NQGSGNIGLVGSYY1175 pKa = 10.4KK1176 pKa = 10.86NSDD1179 pKa = 3.22ADD1181 pKa = 3.74TAAVKK1186 pKa = 10.96VNGFAFQGTVADD1198 pKa = 4.98LGNSANYY1205 pKa = 7.14QTIDD1209 pKa = 2.93IAGAEE1214 pKa = 4.18FNYY1217 pKa = 9.98VHH1219 pKa = 6.08STMNGLAVGNYY1230 pKa = 9.93DD1231 pKa = 3.21SAADD1235 pKa = 3.65HH1236 pKa = 6.6GMGGLPLGPGHH1247 pKa = 7.33AFIYY1251 pKa = 10.35DD1252 pKa = 4.22LNQHH1256 pKa = 6.29KK1257 pKa = 10.0FLTDD1261 pKa = 2.7IMFPGSLSNTAYY1273 pKa = 10.51GIWFNGNDD1281 pKa = 3.62SYY1283 pKa = 10.45TICGGWSPDD1292 pKa = 3.82LVNNLLNQDD1301 pKa = 3.47QPIGKK1306 pKa = 9.9GYY1308 pKa = 10.46LVDD1311 pKa = 4.09YY1312 pKa = 10.36NSKK1315 pKa = 9.12TGAFTHH1321 pKa = 6.25FTSFDD1326 pKa = 3.7YY1327 pKa = 10.96PFGTNFVTHH1336 pKa = 6.46FEE1338 pKa = 4.8GISSVEE1344 pKa = 3.84KK1345 pKa = 10.65GVYY1348 pKa = 8.58TLSADD1353 pKa = 3.65SVQAGSNAPVQGSFVSVLRR1372 pKa = 11.84NTDD1375 pKa = 2.75GSFGSGQWINLNYY1388 pKa = 9.92PGLNPTVNITSSNSVYY1404 pKa = 10.7GNAVVGIVIGANPFSYY1420 pKa = 10.24QATVNVGFQLSNVISGNGGNGIEE1443 pKa = 4.6LDD1445 pKa = 3.77ASNGNQIAMNNVGTDD1460 pKa = 3.5LTGSRR1465 pKa = 11.84DD1466 pKa = 3.34LGNFGNGVLLTGASANNMVGGVATGGNAPTNGVFVRR1502 pKa = 11.84PPEE1505 pKa = 4.36GNLISGNDD1513 pKa = 3.46QNGVLITDD1521 pKa = 4.9LATHH1525 pKa = 6.16NQLSGNFIGTTAPGNSAMGNALDD1548 pKa = 5.78GVAIDD1553 pKa = 4.0NAPGNSLIGCTFQQDD1568 pKa = 3.3PFVFYY1573 pKa = 11.19NVIDD1577 pKa = 4.16GNGGNGLRR1585 pKa = 11.84VTNSDD1590 pKa = 2.82NTTFQANFLGLGADD1604 pKa = 3.27NSTPVGNALDD1614 pKa = 3.94GALINGSSANIQFGGVIPLGNVDD1637 pKa = 3.9AANGRR1642 pKa = 11.84NGVEE1646 pKa = 4.03IADD1649 pKa = 4.01TASGGVYY1656 pKa = 10.45FNTFCGLPAFVDD1668 pKa = 3.71TAVGNTLDD1676 pKa = 3.94GFLVTSTGGNNTIRR1690 pKa = 11.84TNVVSGNKK1698 pKa = 10.01GNGIHH1703 pKa = 7.0ISGNATGVQVTDD1715 pKa = 4.3TIVGLNTSGTTAIPNGANGILLDD1738 pKa = 5.36GNAHH1742 pKa = 6.94DD1743 pKa = 4.49NNIGGFQPSVIPEE1756 pKa = 3.64VTIAANGANGIAVVGNAKK1774 pKa = 10.5NNTIFHH1780 pKa = 6.21SLIGLNISGLTAIGNAGAGIFVGGAATGTIIGGTGPGQKK1819 pKa = 9.98VVVSGNLGGGIQLSGQSQGTQIIGSLIGTNKK1850 pKa = 10.45LGTAPVGNHH1859 pKa = 5.84GNGITIVSSGNSVGGTAAGQGNTVAFNSQFGIVVDD1894 pKa = 4.0TGTGNGILSNSIFNNGSGGILLTTGGNLGQPAPVLTFIPQTGSLTATVNGTLTAVANTTYY1954 pKa = 10.17TVQLFASSSAVPGGQGQFVVSSFSATTNGSGVASFSVTVTIPSSSDD2000 pKa = 3.06QFFTATATDD2009 pKa = 4.14PNKK2012 pKa = 8.72NTSMFSAAVQWTTPNAQYY2030 pKa = 9.04VTSAYY2035 pKa = 10.26QLLLGRR2041 pKa = 11.84MPDD2044 pKa = 3.07ASGGVFWVNQLNAGASPASVVLGIQGSSEE2073 pKa = 4.09YY2074 pKa = 10.34LTDD2077 pKa = 4.28QVFALYY2083 pKa = 10.48GRR2085 pKa = 11.84YY2086 pKa = 9.33LDD2088 pKa = 4.54RR2089 pKa = 11.84APEE2092 pKa = 4.1STGAEE2097 pKa = 3.91FWTNFLIAGGTFEE2110 pKa = 5.17QVAEE2114 pKa = 4.02QLISSPEE2121 pKa = 3.69YY2122 pKa = 10.03FQAHH2126 pKa = 6.71GATNAGFVLGLYY2138 pKa = 10.34ADD2140 pKa = 3.73VFGRR2144 pKa = 11.84TPSRR2148 pKa = 11.84FEE2150 pKa = 3.64LASWVIVLDD2159 pKa = 4.22AGASRR2164 pKa = 11.84LTVATDD2170 pKa = 3.93FLTSLEE2176 pKa = 3.97YY2177 pKa = 10.7RR2178 pKa = 11.84QDD2180 pKa = 3.81LVEE2183 pKa = 4.78SDD2185 pKa = 3.17YY2186 pKa = 11.39DD2187 pKa = 3.44IYY2189 pKa = 11.16LGRR2192 pKa = 11.84TADD2195 pKa = 3.67QNGLNFWVNALNTGSTDD2212 pKa = 3.47QEE2214 pKa = 4.06VLAGIFGSPEE2224 pKa = 3.8GFAKK2228 pKa = 10.46WSS2230 pKa = 3.39
MM1 pKa = 7.13IRR3 pKa = 11.84APRR6 pKa = 11.84TRR8 pKa = 11.84RR9 pKa = 11.84SFNRR13 pKa = 11.84SRR15 pKa = 11.84LWLEE19 pKa = 3.74SLEE22 pKa = 4.27TRR24 pKa = 11.84VTPTTIPVSTLNDD37 pKa = 3.34MGQGSLRR44 pKa = 11.84QAIIDD49 pKa = 3.75ANFNPGPDD57 pKa = 3.47TIEE60 pKa = 3.83FTVAGTIQLTTGALPTITDD79 pKa = 3.61TVKK82 pKa = 10.21IDD84 pKa = 3.23GRR86 pKa = 11.84SAPGFLAAPVVEE98 pKa = 4.49IDD100 pKa = 3.09AHH102 pKa = 5.87GFAGLFFEE110 pKa = 5.09IGSNGSSLLSLAVGNANGAGVTLDD134 pKa = 3.84DD135 pKa = 4.91SNNNVAGNYY144 pKa = 9.32IGLALDD150 pKa = 4.55GSTTASNVGDD160 pKa = 3.42GLVINSSSINNIIGGTSPLDD180 pKa = 3.43RR181 pKa = 11.84NVISGNTGDD190 pKa = 4.86GITVSGSTRR199 pKa = 11.84NQIVGNFIGPDD210 pKa = 3.2ASGLGGRR217 pKa = 11.84GNGLDD222 pKa = 4.91GILLTAGAAVNNVGGTIDD240 pKa = 3.39AQRR243 pKa = 11.84NIISCNAADD252 pKa = 4.56GVSLEE257 pKa = 4.45QGAHH261 pKa = 6.43DD262 pKa = 5.68NIILGNFIGTDD273 pKa = 3.44VTGLAGLGNGLDD285 pKa = 4.0GVHH288 pKa = 7.23ADD290 pKa = 3.75DD291 pKa = 5.29AANNVIGNTVPVKK304 pKa = 10.46NVSYY308 pKa = 10.61FNANLVSPTPNPGWTGIRR326 pKa = 11.84ASDD329 pKa = 3.26TSGNYY334 pKa = 9.86LISGISGTQGLLYY347 pKa = 10.48EE348 pKa = 4.58GTISGAGVTIPVNFPNASASAVYY371 pKa = 10.47GPDD374 pKa = 3.22NLGGNHH380 pKa = 5.27VALVGSYY387 pKa = 10.36NVTGTGLADD396 pKa = 3.13QKK398 pKa = 11.65GFIFEE403 pKa = 4.2GTTADD408 pKa = 3.48LTNPANYY415 pKa = 7.33TTIDD419 pKa = 3.04AGGDD423 pKa = 3.49FNIVHH428 pKa = 6.08STMQGLAVGNSDD440 pKa = 3.63SSPSQGQASLLSHH453 pKa = 6.95AFLYY457 pKa = 10.7DD458 pKa = 3.42VASGQFLPDD467 pKa = 2.86IVFPGSVSNTAYY479 pKa = 10.49GIWFNGGTSYY489 pKa = 9.53TICGGWSSDD498 pKa = 3.41PVNNFDD504 pKa = 4.94DD505 pKa = 3.76QNRR508 pKa = 11.84PIGQGYY514 pKa = 8.82LVDD517 pKa = 4.35YY518 pKa = 11.02DD519 pKa = 4.8SLTQTFTNWTSFQYY533 pKa = 10.73PFGKK537 pKa = 10.28SFFTHH542 pKa = 6.54FEE544 pKa = 4.55GISSVEE550 pKa = 3.87NGVYY554 pKa = 9.5TLNADD559 pKa = 3.72SFQVGTTNPIQGSWVSVRR577 pKa = 11.84RR578 pKa = 11.84NSDD581 pKa = 2.78GSFSLGQWTNLAYY594 pKa = 9.88PGTGNITSSNSVYY607 pKa = 10.13GNQVVGVVFHH617 pKa = 6.86GATAFAFQATVNIGFQLSNVIGCNADD643 pKa = 3.7DD644 pKa = 4.7GVLLTGGATANTIASNFIGLDD665 pKa = 3.49LTGKK669 pKa = 10.14AALANGSDD677 pKa = 3.63GVEE680 pKa = 4.21VEE682 pKa = 4.11NSQGNLIGTTSTGPEE697 pKa = 3.39VNYY700 pKa = 10.31FNTSLITSVPVSGWEE715 pKa = 4.56GIRR718 pKa = 11.84QADD721 pKa = 3.98LPGQFLISGTSGSNGLLFEE740 pKa = 4.19GTIDD744 pKa = 4.62GVGTTNIVDD753 pKa = 3.64YY754 pKa = 9.98PGASSTFVYY763 pKa = 10.62GPDD766 pKa = 3.57NEE768 pKa = 4.73GSGNLGLVGTYY779 pKa = 7.92KK780 pKa = 10.07TANSSVVNGFLFQGTVADD798 pKa = 5.16LGTPANFQTINKK810 pKa = 9.71SGATFNYY817 pKa = 9.54LHH819 pKa = 6.28STMGGLIVGNYY830 pKa = 9.93DD831 pKa = 3.15SAPDD835 pKa = 3.72HH836 pKa = 6.18GQGGLPFGPGHH847 pKa = 5.64ATIYY851 pKa = 10.71DD852 pKa = 3.69IATQTYY858 pKa = 7.89LTDD861 pKa = 3.19IVFPGSKK868 pKa = 9.98SNSVYY873 pKa = 10.87GIWFNGGTSYY883 pKa = 9.4TVCGGYY889 pKa = 10.12SPTFVNNFDD898 pKa = 3.81DD899 pKa = 3.72QNRR902 pKa = 11.84PIGQAYY908 pKa = 9.02LANYY912 pKa = 7.69DD913 pKa = 3.65TATGQFSHH921 pKa = 6.68FTSFSYY927 pKa = 10.7PFGTNFLTHH936 pKa = 6.55FEE938 pKa = 4.92GISSVEE944 pKa = 3.78PGVYY948 pKa = 9.24TLAADD953 pKa = 4.9SIQSGSSQTIQASWVVVRR971 pKa = 11.84RR972 pKa = 11.84NPDD975 pKa = 2.68GSFGTGQWVNLDD987 pKa = 3.67YY988 pKa = 11.2PGVATTGTFTGSDD1001 pKa = 3.25SVYY1004 pKa = 9.18GNQVVGIVVSSTSTFCFQATVNVGTQMSNIISGNSGNGVLLNVGAQNNQVAGNFIGTDD1062 pKa = 3.36ATGSASLGNSLNGVQLEE1079 pKa = 4.37GANNNVIGNNNPITDD1094 pKa = 3.37IPFFNTDD1101 pKa = 3.04SVSVQVSGWQGIRR1114 pKa = 11.84NSDD1117 pKa = 3.3TTGQFLITGTSGTNGLLFDD1136 pKa = 3.87GTIAGVGTSYY1146 pKa = 10.87QVNFPNAATTSVYY1159 pKa = 10.81GPDD1162 pKa = 3.48NQGSGNIGLVGSYY1175 pKa = 10.4KK1176 pKa = 10.86NSDD1179 pKa = 3.22ADD1181 pKa = 3.74TAAVKK1186 pKa = 10.96VNGFAFQGTVADD1198 pKa = 4.98LGNSANYY1205 pKa = 7.14QTIDD1209 pKa = 2.93IAGAEE1214 pKa = 4.18FNYY1217 pKa = 9.98VHH1219 pKa = 6.08STMNGLAVGNYY1230 pKa = 9.93DD1231 pKa = 3.21SAADD1235 pKa = 3.65HH1236 pKa = 6.6GMGGLPLGPGHH1247 pKa = 7.33AFIYY1251 pKa = 10.35DD1252 pKa = 4.22LNQHH1256 pKa = 6.29KK1257 pKa = 10.0FLTDD1261 pKa = 2.7IMFPGSLSNTAYY1273 pKa = 10.51GIWFNGNDD1281 pKa = 3.62SYY1283 pKa = 10.45TICGGWSPDD1292 pKa = 3.82LVNNLLNQDD1301 pKa = 3.47QPIGKK1306 pKa = 9.9GYY1308 pKa = 10.46LVDD1311 pKa = 4.09YY1312 pKa = 10.36NSKK1315 pKa = 9.12TGAFTHH1321 pKa = 6.25FTSFDD1326 pKa = 3.7YY1327 pKa = 10.96PFGTNFVTHH1336 pKa = 6.46FEE1338 pKa = 4.8GISSVEE1344 pKa = 3.84KK1345 pKa = 10.65GVYY1348 pKa = 8.58TLSADD1353 pKa = 3.65SVQAGSNAPVQGSFVSVLRR1372 pKa = 11.84NTDD1375 pKa = 2.75GSFGSGQWINLNYY1388 pKa = 9.92PGLNPTVNITSSNSVYY1404 pKa = 10.7GNAVVGIVIGANPFSYY1420 pKa = 10.24QATVNVGFQLSNVISGNGGNGIEE1443 pKa = 4.6LDD1445 pKa = 3.77ASNGNQIAMNNVGTDD1460 pKa = 3.5LTGSRR1465 pKa = 11.84DD1466 pKa = 3.34LGNFGNGVLLTGASANNMVGGVATGGNAPTNGVFVRR1502 pKa = 11.84PPEE1505 pKa = 4.36GNLISGNDD1513 pKa = 3.46QNGVLITDD1521 pKa = 4.9LATHH1525 pKa = 6.16NQLSGNFIGTTAPGNSAMGNALDD1548 pKa = 5.78GVAIDD1553 pKa = 4.0NAPGNSLIGCTFQQDD1568 pKa = 3.3PFVFYY1573 pKa = 11.19NVIDD1577 pKa = 4.16GNGGNGLRR1585 pKa = 11.84VTNSDD1590 pKa = 2.82NTTFQANFLGLGADD1604 pKa = 3.27NSTPVGNALDD1614 pKa = 3.94GALINGSSANIQFGGVIPLGNVDD1637 pKa = 3.9AANGRR1642 pKa = 11.84NGVEE1646 pKa = 4.03IADD1649 pKa = 4.01TASGGVYY1656 pKa = 10.45FNTFCGLPAFVDD1668 pKa = 3.71TAVGNTLDD1676 pKa = 3.94GFLVTSTGGNNTIRR1690 pKa = 11.84TNVVSGNKK1698 pKa = 10.01GNGIHH1703 pKa = 7.0ISGNATGVQVTDD1715 pKa = 4.3TIVGLNTSGTTAIPNGANGILLDD1738 pKa = 5.36GNAHH1742 pKa = 6.94DD1743 pKa = 4.49NNIGGFQPSVIPEE1756 pKa = 3.64VTIAANGANGIAVVGNAKK1774 pKa = 10.5NNTIFHH1780 pKa = 6.21SLIGLNISGLTAIGNAGAGIFVGGAATGTIIGGTGPGQKK1819 pKa = 9.98VVVSGNLGGGIQLSGQSQGTQIIGSLIGTNKK1850 pKa = 10.45LGTAPVGNHH1859 pKa = 5.84GNGITIVSSGNSVGGTAAGQGNTVAFNSQFGIVVDD1894 pKa = 4.0TGTGNGILSNSIFNNGSGGILLTTGGNLGQPAPVLTFIPQTGSLTATVNGTLTAVANTTYY1954 pKa = 10.17TVQLFASSSAVPGGQGQFVVSSFSATTNGSGVASFSVTVTIPSSSDD2000 pKa = 3.06QFFTATATDD2009 pKa = 4.14PNKK2012 pKa = 8.72NTSMFSAAVQWTTPNAQYY2030 pKa = 9.04VTSAYY2035 pKa = 10.26QLLLGRR2041 pKa = 11.84MPDD2044 pKa = 3.07ASGGVFWVNQLNAGASPASVVLGIQGSSEE2073 pKa = 4.09YY2074 pKa = 10.34LTDD2077 pKa = 4.28QVFALYY2083 pKa = 10.48GRR2085 pKa = 11.84YY2086 pKa = 9.33LDD2088 pKa = 4.54RR2089 pKa = 11.84APEE2092 pKa = 4.1STGAEE2097 pKa = 3.91FWTNFLIAGGTFEE2110 pKa = 5.17QVAEE2114 pKa = 4.02QLISSPEE2121 pKa = 3.69YY2122 pKa = 10.03FQAHH2126 pKa = 6.71GATNAGFVLGLYY2138 pKa = 10.34ADD2140 pKa = 3.73VFGRR2144 pKa = 11.84TPSRR2148 pKa = 11.84FEE2150 pKa = 3.64LASWVIVLDD2159 pKa = 4.22AGASRR2164 pKa = 11.84LTVATDD2170 pKa = 3.93FLTSLEE2176 pKa = 3.97YY2177 pKa = 10.7RR2178 pKa = 11.84QDD2180 pKa = 3.81LVEE2183 pKa = 4.78SDD2185 pKa = 3.17YY2186 pKa = 11.39DD2187 pKa = 3.44IYY2189 pKa = 11.16LGRR2192 pKa = 11.84TADD2195 pKa = 3.67QNGLNFWVNALNTGSTDD2212 pKa = 3.47QEE2214 pKa = 4.06VLAGIFGSPEE2224 pKa = 3.8GFAKK2228 pKa = 10.46WSS2230 pKa = 3.39
Molecular weight: 228.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A090KMN6|A0A090KMN6_9BACT Uncultured bacterium genome assembly Metasoil_fosmids_resub OS=uncultured bacterium OX=77133 PE=4 SV=1
MM1 pKa = 7.88RR2 pKa = 11.84LGPGLSFSWRR12 pKa = 11.84RR13 pKa = 11.84AFGVSAFNARR23 pKa = 11.84IARR26 pKa = 11.84RR27 pKa = 11.84TGIPTTRR34 pKa = 11.84SGLDD38 pKa = 3.03RR39 pKa = 11.84KK40 pKa = 10.06IGRR43 pKa = 11.84FFTAGLLSLLFGGRR57 pKa = 11.84RR58 pKa = 3.39
MM1 pKa = 7.88RR2 pKa = 11.84LGPGLSFSWRR12 pKa = 11.84RR13 pKa = 11.84AFGVSAFNARR23 pKa = 11.84IARR26 pKa = 11.84RR27 pKa = 11.84TGIPTTRR34 pKa = 11.84SGLDD38 pKa = 3.03RR39 pKa = 11.84KK40 pKa = 10.06IGRR43 pKa = 11.84FFTAGLLSLLFGGRR57 pKa = 11.84RR58 pKa = 3.39
Molecular weight: 6.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
357591 |
21 |
2758 |
277.2 |
30.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.039 ± 0.109 | 1.015 ± 0.022 |
5.427 ± 0.041 | 5.61 ± 0.076 |
3.84 ± 0.053 | 8.015 ± 0.103 |
2.232 ± 0.033 | 5.051 ± 0.058 |
3.823 ± 0.062 | 9.965 ± 0.08 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.081 ± 0.033 | 2.986 ± 0.077 |
5.481 ± 0.07 | 3.461 ± 0.048 |
7.521 ± 0.099 | 5.791 ± 0.063 |
5.427 ± 0.068 | 7.235 ± 0.059 |
1.565 ± 0.032 | 2.435 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
