
Hubei tombus-like virus 15
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.64
Get precalculated fractions of proteins
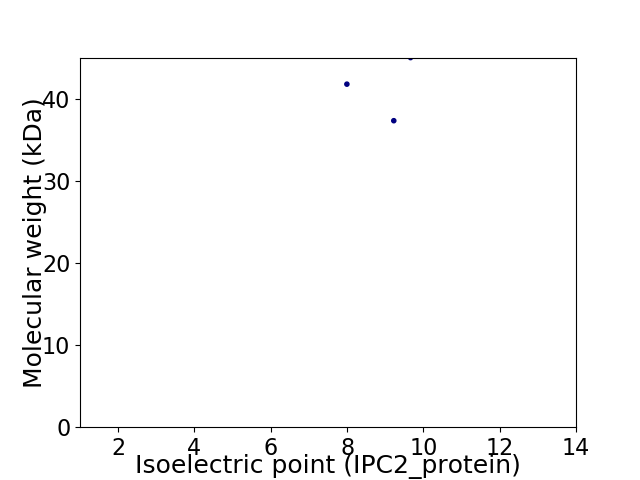
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGT5|A0A1L3KGT5_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 15 OX=1923261 PE=4 SV=1
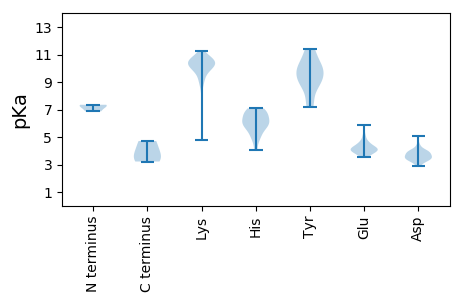
MM1 pKa = 7.27FLKK4 pKa = 10.37ADD6 pKa = 3.78KK7 pKa = 11.04AHH9 pKa = 6.35VFTDD13 pKa = 3.99GNPDD17 pKa = 3.73FGAPRR22 pKa = 11.84CIQYY26 pKa = 10.54RR27 pKa = 11.84NKK29 pKa = 10.43RR30 pKa = 11.84YY31 pKa = 9.56CLRR34 pKa = 11.84LATYY38 pKa = 7.19LHH40 pKa = 5.99PVEE43 pKa = 5.09RR44 pKa = 11.84EE45 pKa = 3.94LYY47 pKa = 9.84KK48 pKa = 11.0KK49 pKa = 10.6LDD51 pKa = 3.41ISGTPIFAKK60 pKa = 10.25GRR62 pKa = 11.84NLVQRR67 pKa = 11.84GQDD70 pKa = 2.87LWAKK74 pKa = 8.71FQHH77 pKa = 5.75FRR79 pKa = 11.84NPTIICMDD87 pKa = 3.57HH88 pKa = 6.72SKK90 pKa = 10.77FDD92 pKa = 3.24AHH94 pKa = 6.88FGVEE98 pKa = 4.12LLRR101 pKa = 11.84LEE103 pKa = 4.29HH104 pKa = 6.82KK105 pKa = 10.07FYY107 pKa = 11.4KK108 pKa = 10.83SMFPKK113 pKa = 10.17CDD115 pKa = 3.4RR116 pKa = 11.84EE117 pKa = 4.06EE118 pKa = 4.09LSRR121 pKa = 11.84LLEE124 pKa = 3.82MQINNKK130 pKa = 9.73GSTKK134 pKa = 10.4HH135 pKa = 4.03GTKK138 pKa = 10.12YY139 pKa = 8.77RR140 pKa = 11.84TRR142 pKa = 11.84GTRR145 pKa = 11.84CSGDD149 pKa = 3.23QNTGMGNSLGNYY161 pKa = 9.7CMLKK165 pKa = 10.67DD166 pKa = 4.03FADD169 pKa = 3.76HH170 pKa = 6.8NGWEE174 pKa = 3.95ACFYY178 pKa = 11.37VDD180 pKa = 5.04GDD182 pKa = 4.01DD183 pKa = 3.62SVMIVEE189 pKa = 5.0GDD191 pKa = 3.81VEE193 pKa = 4.21VDD195 pKa = 3.23PKK197 pKa = 10.89FFKK200 pKa = 10.77QFGMATKK207 pKa = 10.26VDD209 pKa = 3.9VVTKK213 pKa = 9.52EE214 pKa = 3.86FQDD217 pKa = 3.53MEE219 pKa = 4.55FCQTRR224 pKa = 11.84AVFDD228 pKa = 3.95GVSWRR233 pKa = 11.84MVRR236 pKa = 11.84NPARR240 pKa = 11.84LLARR244 pKa = 11.84LPWAVQQVTPQCRR257 pKa = 11.84GKK259 pKa = 9.74YY260 pKa = 9.03LRR262 pKa = 11.84SVGLCEE268 pKa = 3.61IALGVGLPIAQHH280 pKa = 6.18IGEE283 pKa = 4.76KK284 pKa = 10.26LSKK287 pKa = 10.25LGKK290 pKa = 10.28GYY292 pKa = 9.32MVTGNHH298 pKa = 5.06YY299 pKa = 8.12MASKK303 pKa = 10.45EE304 pKa = 4.12YY305 pKa = 10.22IRR307 pKa = 11.84PTRR310 pKa = 11.84ARR312 pKa = 11.84LIEE315 pKa = 4.05PTMEE319 pKa = 3.55ARR321 pKa = 11.84MSYY324 pKa = 9.94ARR326 pKa = 11.84AWGISVEE333 pKa = 4.08EE334 pKa = 4.03QLRR337 pKa = 11.84IEE339 pKa = 4.25TVEE342 pKa = 3.84IEE344 pKa = 4.17LPEE347 pKa = 5.66LGDD350 pKa = 3.76LRR352 pKa = 11.84TFDD355 pKa = 3.8EE356 pKa = 4.59VPYY359 pKa = 9.56RR360 pKa = 11.84TLSS363 pKa = 3.22
MM1 pKa = 7.27FLKK4 pKa = 10.37ADD6 pKa = 3.78KK7 pKa = 11.04AHH9 pKa = 6.35VFTDD13 pKa = 3.99GNPDD17 pKa = 3.73FGAPRR22 pKa = 11.84CIQYY26 pKa = 10.54RR27 pKa = 11.84NKK29 pKa = 10.43RR30 pKa = 11.84YY31 pKa = 9.56CLRR34 pKa = 11.84LATYY38 pKa = 7.19LHH40 pKa = 5.99PVEE43 pKa = 5.09RR44 pKa = 11.84EE45 pKa = 3.94LYY47 pKa = 9.84KK48 pKa = 11.0KK49 pKa = 10.6LDD51 pKa = 3.41ISGTPIFAKK60 pKa = 10.25GRR62 pKa = 11.84NLVQRR67 pKa = 11.84GQDD70 pKa = 2.87LWAKK74 pKa = 8.71FQHH77 pKa = 5.75FRR79 pKa = 11.84NPTIICMDD87 pKa = 3.57HH88 pKa = 6.72SKK90 pKa = 10.77FDD92 pKa = 3.24AHH94 pKa = 6.88FGVEE98 pKa = 4.12LLRR101 pKa = 11.84LEE103 pKa = 4.29HH104 pKa = 6.82KK105 pKa = 10.07FYY107 pKa = 11.4KK108 pKa = 10.83SMFPKK113 pKa = 10.17CDD115 pKa = 3.4RR116 pKa = 11.84EE117 pKa = 4.06EE118 pKa = 4.09LSRR121 pKa = 11.84LLEE124 pKa = 3.82MQINNKK130 pKa = 9.73GSTKK134 pKa = 10.4HH135 pKa = 4.03GTKK138 pKa = 10.12YY139 pKa = 8.77RR140 pKa = 11.84TRR142 pKa = 11.84GTRR145 pKa = 11.84CSGDD149 pKa = 3.23QNTGMGNSLGNYY161 pKa = 9.7CMLKK165 pKa = 10.67DD166 pKa = 4.03FADD169 pKa = 3.76HH170 pKa = 6.8NGWEE174 pKa = 3.95ACFYY178 pKa = 11.37VDD180 pKa = 5.04GDD182 pKa = 4.01DD183 pKa = 3.62SVMIVEE189 pKa = 5.0GDD191 pKa = 3.81VEE193 pKa = 4.21VDD195 pKa = 3.23PKK197 pKa = 10.89FFKK200 pKa = 10.77QFGMATKK207 pKa = 10.26VDD209 pKa = 3.9VVTKK213 pKa = 9.52EE214 pKa = 3.86FQDD217 pKa = 3.53MEE219 pKa = 4.55FCQTRR224 pKa = 11.84AVFDD228 pKa = 3.95GVSWRR233 pKa = 11.84MVRR236 pKa = 11.84NPARR240 pKa = 11.84LLARR244 pKa = 11.84LPWAVQQVTPQCRR257 pKa = 11.84GKK259 pKa = 9.74YY260 pKa = 9.03LRR262 pKa = 11.84SVGLCEE268 pKa = 3.61IALGVGLPIAQHH280 pKa = 6.18IGEE283 pKa = 4.76KK284 pKa = 10.26LSKK287 pKa = 10.25LGKK290 pKa = 10.28GYY292 pKa = 9.32MVTGNHH298 pKa = 5.06YY299 pKa = 8.12MASKK303 pKa = 10.45EE304 pKa = 4.12YY305 pKa = 10.22IRR307 pKa = 11.84PTRR310 pKa = 11.84ARR312 pKa = 11.84LIEE315 pKa = 4.05PTMEE319 pKa = 3.55ARR321 pKa = 11.84MSYY324 pKa = 9.94ARR326 pKa = 11.84AWGISVEE333 pKa = 4.08EE334 pKa = 4.03QLRR337 pKa = 11.84IEE339 pKa = 4.25TVEE342 pKa = 3.84IEE344 pKa = 4.17LPEE347 pKa = 5.66LGDD350 pKa = 3.76LRR352 pKa = 11.84TFDD355 pKa = 3.8EE356 pKa = 4.59VPYY359 pKa = 9.56RR360 pKa = 11.84TLSS363 pKa = 3.22
Molecular weight: 41.81 kDa
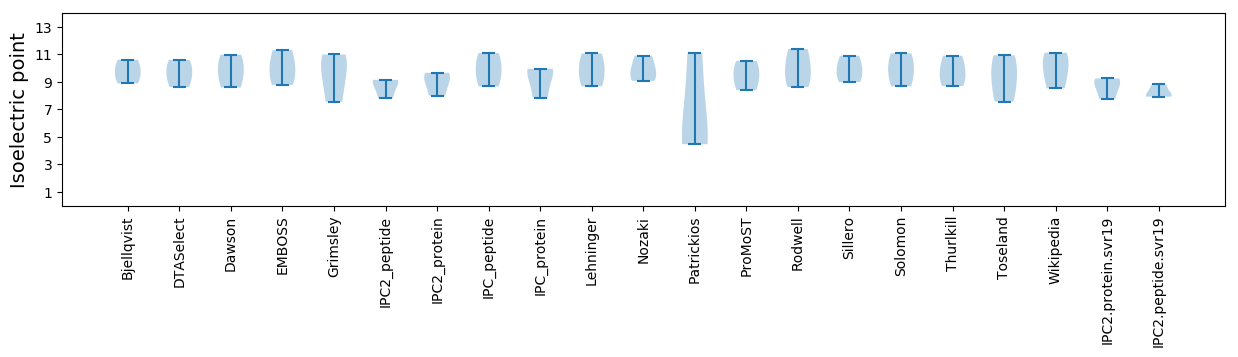
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGX0|A0A1L3KGX0_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 15 OX=1923261 PE=4 SV=1
MM1 pKa = 7.35NKK3 pKa = 9.98SSTGVSTSSLGHH15 pKa = 6.0GKK17 pKa = 10.7GPGQGSHH24 pKa = 5.84RR25 pKa = 11.84VRR27 pKa = 11.84EE28 pKa = 4.15AGANRR33 pKa = 11.84NKK35 pKa = 10.31RR36 pKa = 11.84CSLGVKK42 pKa = 10.0SGGKK46 pKa = 4.82TTNTTGHH53 pKa = 7.08PPAAGWIHH61 pKa = 6.27PQPAANAAAKK71 pKa = 10.01AAATDD76 pKa = 3.97PNKK79 pKa = 10.15IRR81 pKa = 11.84VKK83 pKa = 10.65EE84 pKa = 3.93RR85 pKa = 11.84TTGNGAKK92 pKa = 9.78QGSQQSLLSKK102 pKa = 10.44HH103 pKa = 5.67RR104 pKa = 11.84WSKK107 pKa = 10.48EE108 pKa = 3.63DD109 pKa = 3.38LRR111 pKa = 11.84AAAARR116 pKa = 11.84TLRR119 pKa = 11.84ARR121 pKa = 11.84VKK123 pKa = 9.99QLEE126 pKa = 4.33GEE128 pKa = 4.23LEE130 pKa = 3.95ALRR133 pKa = 11.84KK134 pKa = 9.23QFKK137 pKa = 10.13QSSTGKK143 pKa = 10.31SGGSTPKK150 pKa = 10.57RR151 pKa = 11.84NVDD154 pKa = 3.31KK155 pKa = 10.8PGSKK159 pKa = 10.35VGGKK163 pKa = 8.98QSNQVPKK170 pKa = 10.67SSPPNKK176 pKa = 9.81KK177 pKa = 10.07KK178 pKa = 10.59GDD180 pKa = 3.45RR181 pKa = 11.84RR182 pKa = 11.84TSGRR186 pKa = 11.84PEE188 pKa = 4.15DD189 pKa = 4.07RR190 pKa = 11.84APPAPQEE197 pKa = 3.91QSRR200 pKa = 11.84GDD202 pKa = 3.56RR203 pKa = 11.84TRR205 pKa = 11.84KK206 pKa = 9.39RR207 pKa = 11.84HH208 pKa = 5.11TAPAPPVPPGPSSAMPAKK226 pKa = 10.15VSRR229 pKa = 11.84GTGTAPPAAEE239 pKa = 4.21VTPDD243 pKa = 3.48PAKK246 pKa = 10.58DD247 pKa = 3.43KK248 pKa = 11.26KK249 pKa = 10.54RR250 pKa = 11.84DD251 pKa = 3.34LPQYY255 pKa = 10.35SGPSYY260 pKa = 10.88AATVKK265 pKa = 10.63SGLCTEE271 pKa = 4.28SNKK274 pKa = 10.53PIFSKK279 pKa = 10.67EE280 pKa = 3.57CAIIDD285 pKa = 3.53NSIIARR291 pKa = 11.84SAKK294 pKa = 9.89RR295 pKa = 11.84SMRR298 pKa = 11.84KK299 pKa = 9.05LDD301 pKa = 3.72HH302 pKa = 5.84QWADD306 pKa = 3.0KK307 pKa = 10.81EE308 pKa = 4.36LTHH311 pKa = 6.6FLLMEE316 pKa = 4.38FAFTPRR322 pKa = 11.84TTEE325 pKa = 3.7VLRR328 pKa = 11.84LMHH331 pKa = 6.93GRR333 pKa = 11.84LQKK336 pKa = 10.42HH337 pKa = 5.96LRR339 pKa = 11.84TFDD342 pKa = 3.09TSAYY346 pKa = 7.26TQEE349 pKa = 4.0EE350 pKa = 5.2MYY352 pKa = 10.07KK353 pKa = 10.43LSVRR357 pKa = 11.84TVEE360 pKa = 3.78AAIRR364 pKa = 11.84VPQAEE369 pKa = 4.09QAVRR373 pKa = 11.84AGLKK377 pKa = 9.91NDD379 pKa = 3.43SALEE383 pKa = 3.9EE384 pKa = 3.95VRR386 pKa = 11.84KK387 pKa = 10.21NNKK390 pKa = 9.12FLEE393 pKa = 4.66SGLVGNVRR401 pKa = 11.84GLLGNKK407 pKa = 8.79KK408 pKa = 9.84LKK410 pKa = 10.28EE411 pKa = 4.1MPGKK415 pKa = 10.51SRR417 pKa = 3.7
MM1 pKa = 7.35NKK3 pKa = 9.98SSTGVSTSSLGHH15 pKa = 6.0GKK17 pKa = 10.7GPGQGSHH24 pKa = 5.84RR25 pKa = 11.84VRR27 pKa = 11.84EE28 pKa = 4.15AGANRR33 pKa = 11.84NKK35 pKa = 10.31RR36 pKa = 11.84CSLGVKK42 pKa = 10.0SGGKK46 pKa = 4.82TTNTTGHH53 pKa = 7.08PPAAGWIHH61 pKa = 6.27PQPAANAAAKK71 pKa = 10.01AAATDD76 pKa = 3.97PNKK79 pKa = 10.15IRR81 pKa = 11.84VKK83 pKa = 10.65EE84 pKa = 3.93RR85 pKa = 11.84TTGNGAKK92 pKa = 9.78QGSQQSLLSKK102 pKa = 10.44HH103 pKa = 5.67RR104 pKa = 11.84WSKK107 pKa = 10.48EE108 pKa = 3.63DD109 pKa = 3.38LRR111 pKa = 11.84AAAARR116 pKa = 11.84TLRR119 pKa = 11.84ARR121 pKa = 11.84VKK123 pKa = 9.99QLEE126 pKa = 4.33GEE128 pKa = 4.23LEE130 pKa = 3.95ALRR133 pKa = 11.84KK134 pKa = 9.23QFKK137 pKa = 10.13QSSTGKK143 pKa = 10.31SGGSTPKK150 pKa = 10.57RR151 pKa = 11.84NVDD154 pKa = 3.31KK155 pKa = 10.8PGSKK159 pKa = 10.35VGGKK163 pKa = 8.98QSNQVPKK170 pKa = 10.67SSPPNKK176 pKa = 9.81KK177 pKa = 10.07KK178 pKa = 10.59GDD180 pKa = 3.45RR181 pKa = 11.84RR182 pKa = 11.84TSGRR186 pKa = 11.84PEE188 pKa = 4.15DD189 pKa = 4.07RR190 pKa = 11.84APPAPQEE197 pKa = 3.91QSRR200 pKa = 11.84GDD202 pKa = 3.56RR203 pKa = 11.84TRR205 pKa = 11.84KK206 pKa = 9.39RR207 pKa = 11.84HH208 pKa = 5.11TAPAPPVPPGPSSAMPAKK226 pKa = 10.15VSRR229 pKa = 11.84GTGTAPPAAEE239 pKa = 4.21VTPDD243 pKa = 3.48PAKK246 pKa = 10.58DD247 pKa = 3.43KK248 pKa = 11.26KK249 pKa = 10.54RR250 pKa = 11.84DD251 pKa = 3.34LPQYY255 pKa = 10.35SGPSYY260 pKa = 10.88AATVKK265 pKa = 10.63SGLCTEE271 pKa = 4.28SNKK274 pKa = 10.53PIFSKK279 pKa = 10.67EE280 pKa = 3.57CAIIDD285 pKa = 3.53NSIIARR291 pKa = 11.84SAKK294 pKa = 9.89RR295 pKa = 11.84SMRR298 pKa = 11.84KK299 pKa = 9.05LDD301 pKa = 3.72HH302 pKa = 5.84QWADD306 pKa = 3.0KK307 pKa = 10.81EE308 pKa = 4.36LTHH311 pKa = 6.6FLLMEE316 pKa = 4.38FAFTPRR322 pKa = 11.84TTEE325 pKa = 3.7VLRR328 pKa = 11.84LMHH331 pKa = 6.93GRR333 pKa = 11.84LQKK336 pKa = 10.42HH337 pKa = 5.96LRR339 pKa = 11.84TFDD342 pKa = 3.09TSAYY346 pKa = 7.26TQEE349 pKa = 4.0EE350 pKa = 5.2MYY352 pKa = 10.07KK353 pKa = 10.43LSVRR357 pKa = 11.84TVEE360 pKa = 3.78AAIRR364 pKa = 11.84VPQAEE369 pKa = 4.09QAVRR373 pKa = 11.84AGLKK377 pKa = 9.91NDD379 pKa = 3.43SALEE383 pKa = 3.9EE384 pKa = 3.95VRR386 pKa = 11.84KK387 pKa = 10.21NNKK390 pKa = 9.12FLEE393 pKa = 4.66SGLVGNVRR401 pKa = 11.84GLLGNKK407 pKa = 8.79KK408 pKa = 9.84LKK410 pKa = 10.28EE411 pKa = 4.1MPGKK415 pKa = 10.51SRR417 pKa = 3.7
Molecular weight: 45.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1129 |
349 |
417 |
376.3 |
41.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.149 ± 1.039 | 1.594 ± 0.45 |
3.809 ± 0.7 | 5.049 ± 0.725 |
2.923 ± 0.797 | 8.415 ± 0.255 |
1.86 ± 0.597 | 3.012 ± 0.518 |
6.289 ± 2.352 | 7.44 ± 0.469 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.48 ± 0.482 | 4.34 ± 0.525 |
6.377 ± 0.785 | 4.074 ± 0.113 |
7.972 ± 0.378 | 7.352 ± 1.113 |
8.06 ± 1.716 | 6.997 ± 1.417 |
1.417 ± 0.404 | 2.391 ± 0.679 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
