
Phodopus sungorus papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Pipapillomavirus; Pipapillomavirus 1
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
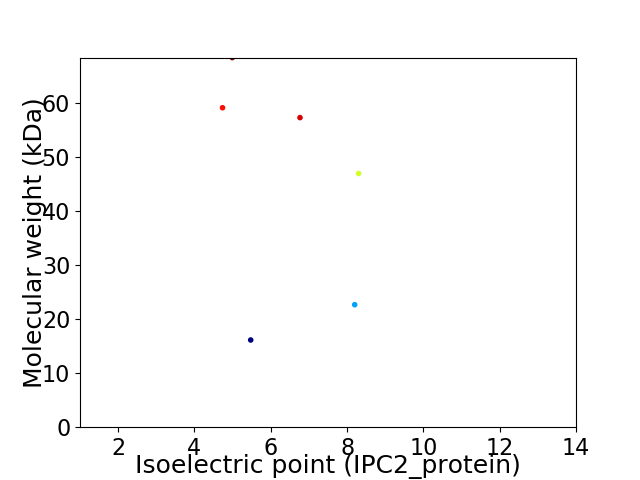
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W8ZM63|W8ZM63_9PAPI Minor capsid protein L2 OS=Phodopus sungorus papillomavirus 1 OX=1487796 GN=L2 PE=3 SV=1
MM1 pKa = 6.93VAVNRR6 pKa = 11.84ARR8 pKa = 11.84RR9 pKa = 11.84TKK11 pKa = 10.33RR12 pKa = 11.84DD13 pKa = 3.37SASNLYY19 pKa = 9.65RR20 pKa = 11.84QCQVTGNCPPDD31 pKa = 3.52VVNKK35 pKa = 10.44IEE37 pKa = 4.09GTTLADD43 pKa = 3.4KK44 pKa = 10.55LSKK47 pKa = 10.35IFASILYY54 pKa = 10.19LGGLGIGTGRR64 pKa = 11.84GSGGATGYY72 pKa = 11.27GPINPGGGRR81 pKa = 11.84ITGTGTVMRR90 pKa = 11.84PGVVIDD96 pKa = 3.83PVGPNDD102 pKa = 4.26IITVGATDD110 pKa = 3.61SSIVPLLEE118 pKa = 4.09ATPDD122 pKa = 3.28IPIEE126 pKa = 4.48GGPEE130 pKa = 3.79VPPAGPDD137 pKa = 3.19VSTVDD142 pKa = 3.03VTANVDD148 pKa = 3.94PISEE152 pKa = 4.45VNVTTGSTITNPDD165 pKa = 3.21SAVIDD170 pKa = 3.93VQPAPSGPRR179 pKa = 11.84RR180 pKa = 11.84VTVSRR185 pKa = 11.84SEE187 pKa = 4.07FQNASYY193 pKa = 11.39VSVTHH198 pKa = 6.8PSQGLGEE205 pKa = 4.28SGGALVSADD214 pKa = 3.26SSGSFVSSGHH224 pKa = 5.83EE225 pKa = 3.76LDD227 pKa = 3.11IGIIIGEE234 pKa = 4.26RR235 pKa = 11.84PLGEE239 pKa = 4.35TNFEE243 pKa = 4.38AIEE246 pKa = 4.15LDD248 pKa = 4.01EE249 pKa = 4.52ILTTGGTAEE258 pKa = 3.92FDD260 pKa = 2.87IGEE263 pKa = 4.4GVNRR267 pKa = 11.84VGPSTSTPDD276 pKa = 3.27NYY278 pKa = 10.8LGRR281 pKa = 11.84ALSRR285 pKa = 11.84FRR287 pKa = 11.84EE288 pKa = 4.33IYY290 pKa = 10.21DD291 pKa = 3.79RR292 pKa = 11.84PRR294 pKa = 11.84DD295 pKa = 3.53LYY297 pKa = 10.87NRR299 pKa = 11.84RR300 pKa = 11.84VQQVRR305 pKa = 11.84VRR307 pKa = 11.84NRR309 pKa = 11.84DD310 pKa = 3.14QFLGAPGRR318 pKa = 11.84LVTYY322 pKa = 10.05EE323 pKa = 3.83FDD325 pKa = 3.55NPAFAGVEE333 pKa = 4.2DD334 pKa = 4.49EE335 pKa = 4.52VSLIFQQDD343 pKa = 3.57LNEE346 pKa = 4.2VQAAPDD352 pKa = 3.94TDD354 pKa = 3.78FMDD357 pKa = 5.11IIRR360 pKa = 11.84LGRR363 pKa = 11.84EE364 pKa = 4.31RR365 pKa = 11.84IAEE368 pKa = 4.21TADD371 pKa = 3.05GTVRR375 pKa = 11.84ISRR378 pKa = 11.84LGQRR382 pKa = 11.84GTIRR386 pKa = 11.84TRR388 pKa = 11.84SGLQIGGKK396 pKa = 6.93VHH398 pKa = 7.5FYY400 pKa = 10.96TDD402 pKa = 4.01LSPIATEE409 pKa = 4.24NIEE412 pKa = 4.3LSTLGEE418 pKa = 4.11VSGEE422 pKa = 4.14SMTIDD427 pKa = 3.39ALNEE431 pKa = 3.57YY432 pKa = 10.67SLISEE437 pKa = 4.41SGIEE441 pKa = 4.13PVPFPDD447 pKa = 3.98EE448 pKa = 4.79DD449 pKa = 3.96LLDD452 pKa = 3.61VHH454 pKa = 8.26SEE456 pKa = 4.04DD457 pKa = 4.2FSGSRR462 pKa = 11.84LHH464 pKa = 6.85IFRR467 pKa = 11.84GGNTRR472 pKa = 11.84FGVVYY477 pKa = 9.91EE478 pKa = 4.19PSEE481 pKa = 4.18SLGARR486 pKa = 11.84TIFPDD491 pKa = 3.08IDD493 pKa = 3.2TGDD496 pKa = 3.8FVRR499 pKa = 11.84YY500 pKa = 7.93PQTNVSPADD509 pKa = 3.26IPGRR513 pKa = 11.84DD514 pKa = 4.29LVPVDD519 pKa = 4.27PGTTHH524 pKa = 6.67IVGSEE529 pKa = 4.04FSSVDD534 pKa = 3.67YY535 pKa = 10.65YY536 pKa = 11.16LHH538 pKa = 6.68PSLRR542 pKa = 11.84RR543 pKa = 11.84RR544 pKa = 11.84KK545 pKa = 9.57RR546 pKa = 11.84KK547 pKa = 10.03RR548 pKa = 11.84NFHH551 pKa = 6.21
MM1 pKa = 6.93VAVNRR6 pKa = 11.84ARR8 pKa = 11.84RR9 pKa = 11.84TKK11 pKa = 10.33RR12 pKa = 11.84DD13 pKa = 3.37SASNLYY19 pKa = 9.65RR20 pKa = 11.84QCQVTGNCPPDD31 pKa = 3.52VVNKK35 pKa = 10.44IEE37 pKa = 4.09GTTLADD43 pKa = 3.4KK44 pKa = 10.55LSKK47 pKa = 10.35IFASILYY54 pKa = 10.19LGGLGIGTGRR64 pKa = 11.84GSGGATGYY72 pKa = 11.27GPINPGGGRR81 pKa = 11.84ITGTGTVMRR90 pKa = 11.84PGVVIDD96 pKa = 3.83PVGPNDD102 pKa = 4.26IITVGATDD110 pKa = 3.61SSIVPLLEE118 pKa = 4.09ATPDD122 pKa = 3.28IPIEE126 pKa = 4.48GGPEE130 pKa = 3.79VPPAGPDD137 pKa = 3.19VSTVDD142 pKa = 3.03VTANVDD148 pKa = 3.94PISEE152 pKa = 4.45VNVTTGSTITNPDD165 pKa = 3.21SAVIDD170 pKa = 3.93VQPAPSGPRR179 pKa = 11.84RR180 pKa = 11.84VTVSRR185 pKa = 11.84SEE187 pKa = 4.07FQNASYY193 pKa = 11.39VSVTHH198 pKa = 6.8PSQGLGEE205 pKa = 4.28SGGALVSADD214 pKa = 3.26SSGSFVSSGHH224 pKa = 5.83EE225 pKa = 3.76LDD227 pKa = 3.11IGIIIGEE234 pKa = 4.26RR235 pKa = 11.84PLGEE239 pKa = 4.35TNFEE243 pKa = 4.38AIEE246 pKa = 4.15LDD248 pKa = 4.01EE249 pKa = 4.52ILTTGGTAEE258 pKa = 3.92FDD260 pKa = 2.87IGEE263 pKa = 4.4GVNRR267 pKa = 11.84VGPSTSTPDD276 pKa = 3.27NYY278 pKa = 10.8LGRR281 pKa = 11.84ALSRR285 pKa = 11.84FRR287 pKa = 11.84EE288 pKa = 4.33IYY290 pKa = 10.21DD291 pKa = 3.79RR292 pKa = 11.84PRR294 pKa = 11.84DD295 pKa = 3.53LYY297 pKa = 10.87NRR299 pKa = 11.84RR300 pKa = 11.84VQQVRR305 pKa = 11.84VRR307 pKa = 11.84NRR309 pKa = 11.84DD310 pKa = 3.14QFLGAPGRR318 pKa = 11.84LVTYY322 pKa = 10.05EE323 pKa = 3.83FDD325 pKa = 3.55NPAFAGVEE333 pKa = 4.2DD334 pKa = 4.49EE335 pKa = 4.52VSLIFQQDD343 pKa = 3.57LNEE346 pKa = 4.2VQAAPDD352 pKa = 3.94TDD354 pKa = 3.78FMDD357 pKa = 5.11IIRR360 pKa = 11.84LGRR363 pKa = 11.84EE364 pKa = 4.31RR365 pKa = 11.84IAEE368 pKa = 4.21TADD371 pKa = 3.05GTVRR375 pKa = 11.84ISRR378 pKa = 11.84LGQRR382 pKa = 11.84GTIRR386 pKa = 11.84TRR388 pKa = 11.84SGLQIGGKK396 pKa = 6.93VHH398 pKa = 7.5FYY400 pKa = 10.96TDD402 pKa = 4.01LSPIATEE409 pKa = 4.24NIEE412 pKa = 4.3LSTLGEE418 pKa = 4.11VSGEE422 pKa = 4.14SMTIDD427 pKa = 3.39ALNEE431 pKa = 3.57YY432 pKa = 10.67SLISEE437 pKa = 4.41SGIEE441 pKa = 4.13PVPFPDD447 pKa = 3.98EE448 pKa = 4.79DD449 pKa = 3.96LLDD452 pKa = 3.61VHH454 pKa = 8.26SEE456 pKa = 4.04DD457 pKa = 4.2FSGSRR462 pKa = 11.84LHH464 pKa = 6.85IFRR467 pKa = 11.84GGNTRR472 pKa = 11.84FGVVYY477 pKa = 9.91EE478 pKa = 4.19PSEE481 pKa = 4.18SLGARR486 pKa = 11.84TIFPDD491 pKa = 3.08IDD493 pKa = 3.2TGDD496 pKa = 3.8FVRR499 pKa = 11.84YY500 pKa = 7.93PQTNVSPADD509 pKa = 3.26IPGRR513 pKa = 11.84DD514 pKa = 4.29LVPVDD519 pKa = 4.27PGTTHH524 pKa = 6.67IVGSEE529 pKa = 4.04FSSVDD534 pKa = 3.67YY535 pKa = 10.65YY536 pKa = 11.16LHH538 pKa = 6.68PSLRR542 pKa = 11.84RR543 pKa = 11.84RR544 pKa = 11.84KK545 pKa = 9.57RR546 pKa = 11.84KK547 pKa = 10.03RR548 pKa = 11.84NFHH551 pKa = 6.21
Molecular weight: 59.07 kDa
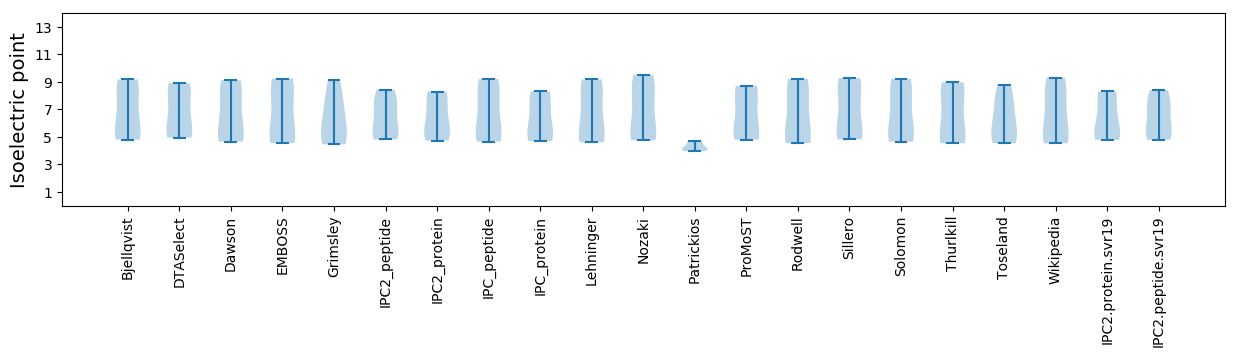
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A078BR99|A0A078BR99_9PAPI Protein E7 OS=Phodopus sungorus papillomavirus 1 OX=1487796 GN=E7 PE=3 SV=1
MM1 pKa = 7.24PTLTEE6 pKa = 4.35RR7 pKa = 11.84FDD9 pKa = 3.95AVQDD13 pKa = 3.86QILSLIEE20 pKa = 4.24RR21 pKa = 11.84GSTRR25 pKa = 11.84LADD28 pKa = 3.54HH29 pKa = 5.81VVYY32 pKa = 10.22WDD34 pKa = 3.66LVRR37 pKa = 11.84KK38 pKa = 9.95EE39 pKa = 4.86GVLQYY44 pKa = 9.69YY45 pKa = 10.49ARR47 pKa = 11.84RR48 pKa = 11.84QNMTRR53 pKa = 11.84LGLLPLPSQLGAEE66 pKa = 4.56HH67 pKa = 7.04KK68 pKa = 10.33AKK70 pKa = 10.5KK71 pKa = 9.45AIQMGLLLRR80 pKa = 11.84SLAEE84 pKa = 3.93SPFANEE90 pKa = 3.43TWTVTDD96 pKa = 3.4TSVEE100 pKa = 4.31VYY102 pKa = 10.14EE103 pKa = 4.13QTDD106 pKa = 3.54PEE108 pKa = 4.4KK109 pKa = 10.22TFKK112 pKa = 11.1KK113 pKa = 9.98NGQTIEE119 pKa = 3.73VWYY122 pKa = 10.42DD123 pKa = 3.19NDD125 pKa = 4.01PEE127 pKa = 4.25NSVAYY132 pKa = 7.8TLWRR136 pKa = 11.84YY137 pKa = 9.14IYY139 pKa = 10.41KK140 pKa = 9.86QDD142 pKa = 5.4EE143 pKa = 3.87NGQWHH148 pKa = 6.59KK149 pKa = 11.53LEE151 pKa = 4.26GRR153 pKa = 11.84VDD155 pKa = 3.72YY156 pKa = 11.32AGLYY160 pKa = 10.07YY161 pKa = 9.72IDD163 pKa = 4.19PSGMRR168 pKa = 11.84VYY170 pKa = 11.15YY171 pKa = 10.55EE172 pKa = 5.36DD173 pKa = 4.02FFEE176 pKa = 5.03DD177 pKa = 3.46ADD179 pKa = 4.53RR180 pKa = 11.84YY181 pKa = 9.45GHH183 pKa = 6.32SSTWTVKK190 pKa = 10.18GDD192 pKa = 3.51KK193 pKa = 10.39DD194 pKa = 3.02ISAPVTSSDD203 pKa = 3.47KK204 pKa = 10.44SWRR207 pKa = 11.84AQEE210 pKa = 4.18EE211 pKa = 4.22QPKK214 pKa = 8.78TKK216 pKa = 9.98RR217 pKa = 11.84RR218 pKa = 11.84RR219 pKa = 11.84DD220 pKa = 3.47NPTTSTPEE228 pKa = 3.14WDD230 pKa = 3.72YY231 pKa = 11.67PEE233 pKa = 4.53TSEE236 pKa = 5.17QEE238 pKa = 4.06EE239 pKa = 4.5RR240 pKa = 11.84PQTSTDD246 pKa = 3.05TPRR249 pKa = 11.84VKK251 pKa = 10.34RR252 pKa = 11.84QRR254 pKa = 11.84QADD257 pKa = 3.92SPSGRR262 pKa = 11.84RR263 pKa = 11.84GRR265 pKa = 11.84PWSEE269 pKa = 3.49STPRR273 pKa = 11.84NGRR276 pKa = 11.84GGRR279 pKa = 11.84EE280 pKa = 3.81QRR282 pKa = 11.84EE283 pKa = 4.21PSPDD287 pKa = 3.18TSPINGGGLSYY298 pKa = 10.95DD299 pKa = 3.86QIGSSHH305 pKa = 4.67TTVPRR310 pKa = 11.84RR311 pKa = 11.84GIGRR315 pKa = 11.84GQRR318 pKa = 11.84LLLEE322 pKa = 4.64ARR324 pKa = 11.84DD325 pKa = 3.92PPVILITGPANSLKK339 pKa = 10.12CWRR342 pKa = 11.84NRR344 pKa = 11.84VRR346 pKa = 11.84RR347 pKa = 11.84RR348 pKa = 11.84PVRR351 pKa = 11.84LFDD354 pKa = 5.45RR355 pKa = 11.84ISTCFSWVTEE365 pKa = 4.02GSGDD369 pKa = 3.5GAGTQRR375 pKa = 11.84LLIAFKK381 pKa = 11.02DD382 pKa = 3.55EE383 pKa = 4.2TQRR386 pKa = 11.84EE387 pKa = 4.5LFLKK391 pKa = 8.46TVPLPRR397 pKa = 11.84GSKK400 pKa = 9.77FFKK403 pKa = 10.94GNLDD407 pKa = 3.63GLL409 pKa = 4.27
MM1 pKa = 7.24PTLTEE6 pKa = 4.35RR7 pKa = 11.84FDD9 pKa = 3.95AVQDD13 pKa = 3.86QILSLIEE20 pKa = 4.24RR21 pKa = 11.84GSTRR25 pKa = 11.84LADD28 pKa = 3.54HH29 pKa = 5.81VVYY32 pKa = 10.22WDD34 pKa = 3.66LVRR37 pKa = 11.84KK38 pKa = 9.95EE39 pKa = 4.86GVLQYY44 pKa = 9.69YY45 pKa = 10.49ARR47 pKa = 11.84RR48 pKa = 11.84QNMTRR53 pKa = 11.84LGLLPLPSQLGAEE66 pKa = 4.56HH67 pKa = 7.04KK68 pKa = 10.33AKK70 pKa = 10.5KK71 pKa = 9.45AIQMGLLLRR80 pKa = 11.84SLAEE84 pKa = 3.93SPFANEE90 pKa = 3.43TWTVTDD96 pKa = 3.4TSVEE100 pKa = 4.31VYY102 pKa = 10.14EE103 pKa = 4.13QTDD106 pKa = 3.54PEE108 pKa = 4.4KK109 pKa = 10.22TFKK112 pKa = 11.1KK113 pKa = 9.98NGQTIEE119 pKa = 3.73VWYY122 pKa = 10.42DD123 pKa = 3.19NDD125 pKa = 4.01PEE127 pKa = 4.25NSVAYY132 pKa = 7.8TLWRR136 pKa = 11.84YY137 pKa = 9.14IYY139 pKa = 10.41KK140 pKa = 9.86QDD142 pKa = 5.4EE143 pKa = 3.87NGQWHH148 pKa = 6.59KK149 pKa = 11.53LEE151 pKa = 4.26GRR153 pKa = 11.84VDD155 pKa = 3.72YY156 pKa = 11.32AGLYY160 pKa = 10.07YY161 pKa = 9.72IDD163 pKa = 4.19PSGMRR168 pKa = 11.84VYY170 pKa = 11.15YY171 pKa = 10.55EE172 pKa = 5.36DD173 pKa = 4.02FFEE176 pKa = 5.03DD177 pKa = 3.46ADD179 pKa = 4.53RR180 pKa = 11.84YY181 pKa = 9.45GHH183 pKa = 6.32SSTWTVKK190 pKa = 10.18GDD192 pKa = 3.51KK193 pKa = 10.39DD194 pKa = 3.02ISAPVTSSDD203 pKa = 3.47KK204 pKa = 10.44SWRR207 pKa = 11.84AQEE210 pKa = 4.18EE211 pKa = 4.22QPKK214 pKa = 8.78TKK216 pKa = 9.98RR217 pKa = 11.84RR218 pKa = 11.84RR219 pKa = 11.84DD220 pKa = 3.47NPTTSTPEE228 pKa = 3.14WDD230 pKa = 3.72YY231 pKa = 11.67PEE233 pKa = 4.53TSEE236 pKa = 5.17QEE238 pKa = 4.06EE239 pKa = 4.5RR240 pKa = 11.84PQTSTDD246 pKa = 3.05TPRR249 pKa = 11.84VKK251 pKa = 10.34RR252 pKa = 11.84QRR254 pKa = 11.84QADD257 pKa = 3.92SPSGRR262 pKa = 11.84RR263 pKa = 11.84GRR265 pKa = 11.84PWSEE269 pKa = 3.49STPRR273 pKa = 11.84NGRR276 pKa = 11.84GGRR279 pKa = 11.84EE280 pKa = 3.81QRR282 pKa = 11.84EE283 pKa = 4.21PSPDD287 pKa = 3.18TSPINGGGLSYY298 pKa = 10.95DD299 pKa = 3.86QIGSSHH305 pKa = 4.67TTVPRR310 pKa = 11.84RR311 pKa = 11.84GIGRR315 pKa = 11.84GQRR318 pKa = 11.84LLLEE322 pKa = 4.64ARR324 pKa = 11.84DD325 pKa = 3.92PPVILITGPANSLKK339 pKa = 10.12CWRR342 pKa = 11.84NRR344 pKa = 11.84VRR346 pKa = 11.84RR347 pKa = 11.84RR348 pKa = 11.84PVRR351 pKa = 11.84LFDD354 pKa = 5.45RR355 pKa = 11.84ISTCFSWVTEE365 pKa = 4.02GSGDD369 pKa = 3.5GAGTQRR375 pKa = 11.84LLIAFKK381 pKa = 11.02DD382 pKa = 3.55EE383 pKa = 4.2TQRR386 pKa = 11.84EE387 pKa = 4.5LFLKK391 pKa = 8.46TVPLPRR397 pKa = 11.84GSKK400 pKa = 9.77FFKK403 pKa = 10.94GNLDD407 pKa = 3.63GLL409 pKa = 4.27
Molecular weight: 46.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2415 |
145 |
609 |
402.5 |
45.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.128 ± 0.786 | 1.988 ± 0.648 |
6.708 ± 0.373 | 6.087 ± 0.387 |
4.224 ± 0.48 | 7.122 ± 1.157 |
1.449 ± 0.101 | 4.679 ± 0.86 |
4.596 ± 0.875 | 8.737 ± 0.765 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.781 ± 0.411 | 3.892 ± 0.605 |
6.377 ± 0.784 | 4.555 ± 0.58 |
7.039 ± 0.79 | 7.329 ± 0.96 |
6.211 ± 0.578 | 6.708 ± 0.632 |
1.284 ± 0.408 | 3.106 ± 0.223 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
