
Cyanoramphus nest associated circular X DNA virus
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.47
Get precalculated fractions of proteins
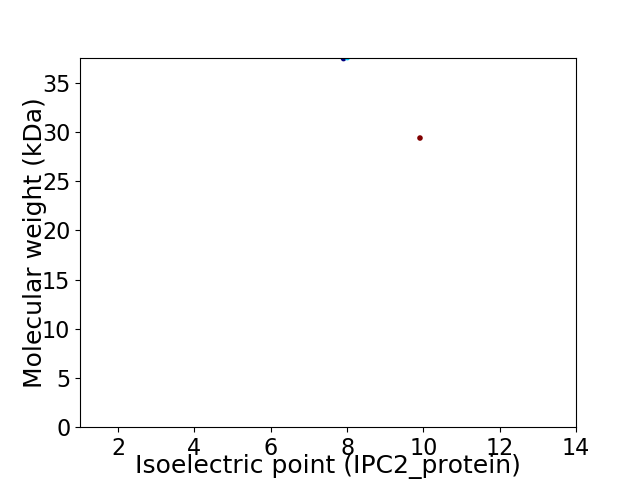
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L7UX06|L7UX06_9VIRU Putative capsid protein OS=Cyanoramphus nest associated circular X DNA virus OX=1282443 PE=4 SV=1
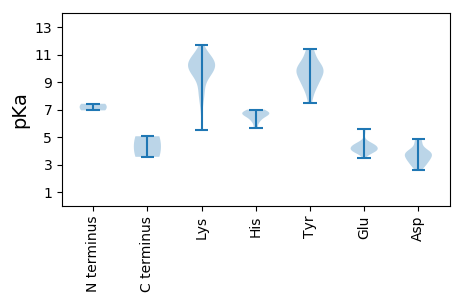
MM1 pKa = 7.42NFQRR5 pKa = 11.84KK6 pKa = 7.6RR7 pKa = 11.84WCFTVNNYY15 pKa = 8.82TEE17 pKa = 5.61DD18 pKa = 3.54EE19 pKa = 4.51FNSIKK24 pKa = 10.64QNFCEE29 pKa = 4.14RR30 pKa = 11.84VAKK33 pKa = 9.38FAVMGRR39 pKa = 11.84EE40 pKa = 3.95IGSGGVPHH48 pKa = 6.74LQGFISLQNKK58 pKa = 8.17VRR60 pKa = 11.84LTGIKK65 pKa = 10.22AIVGQRR71 pKa = 11.84AHH73 pKa = 6.62VEE75 pKa = 3.97PAKK78 pKa = 10.45GTDD81 pKa = 3.37QKK83 pKa = 11.66NLEE86 pKa = 4.5YY87 pKa = 9.48CTKK90 pKa = 10.5SCGVDD95 pKa = 2.84GATIYY100 pKa = 10.07GVPAGGVTEE109 pKa = 4.57KK110 pKa = 11.23GGGTPLSRR118 pKa = 11.84RR119 pKa = 11.84ALEE122 pKa = 4.22CVRR125 pKa = 11.84KK126 pKa = 9.92RR127 pKa = 11.84SAGTSISEE135 pKa = 4.65LLDD138 pKa = 4.63DD139 pKa = 4.84DD140 pKa = 4.72TLAPAYY146 pKa = 9.14IVHH149 pKa = 6.28KK150 pKa = 10.49RR151 pKa = 11.84AIEE154 pKa = 3.99EE155 pKa = 4.31CASDD159 pKa = 3.45IAGAQQFQAEE169 pKa = 4.41RR170 pKa = 11.84ARR172 pKa = 11.84YY173 pKa = 7.45ATTRR177 pKa = 11.84WKK179 pKa = 10.24PFQFNILKK187 pKa = 9.78ICMQEE192 pKa = 3.51PHH194 pKa = 6.78PRR196 pKa = 11.84AVHH199 pKa = 5.65WFWDD203 pKa = 3.69AEE205 pKa = 4.35GNTGKK210 pKa = 8.27TFIAKK215 pKa = 10.05YY216 pKa = 10.3LVLLHH221 pKa = 6.8DD222 pKa = 4.37AVRR225 pKa = 11.84FEE227 pKa = 4.45NGKK230 pKa = 9.75SADD233 pKa = 2.76IKK235 pKa = 10.72YY236 pKa = 10.06AYY238 pKa = 9.1KK239 pKa = 10.15GQRR242 pKa = 11.84IVIFDD247 pKa = 3.87FSRR250 pKa = 11.84SQTDD254 pKa = 3.58HH255 pKa = 6.52INYY258 pKa = 9.98EE259 pKa = 3.95VMEE262 pKa = 4.34SVKK265 pKa = 10.85NGIVFSPKK273 pKa = 9.53YY274 pKa = 8.59EE275 pKa = 4.0SGMKK279 pKa = 9.92VFRR282 pKa = 11.84TPHH285 pKa = 6.14MICFANEE292 pKa = 3.92RR293 pKa = 11.84PDD295 pKa = 3.76TSNMSMDD302 pKa = 2.64RR303 pKa = 11.84WEE305 pKa = 3.73IHH307 pKa = 6.99EE308 pKa = 4.04ITTRR312 pKa = 11.84QARR315 pKa = 11.84TPTTLPDD322 pKa = 3.88RR323 pKa = 11.84DD324 pKa = 3.87SDD326 pKa = 3.79THH328 pKa = 6.7EE329 pKa = 4.01IAA331 pKa = 5.06
MM1 pKa = 7.42NFQRR5 pKa = 11.84KK6 pKa = 7.6RR7 pKa = 11.84WCFTVNNYY15 pKa = 8.82TEE17 pKa = 5.61DD18 pKa = 3.54EE19 pKa = 4.51FNSIKK24 pKa = 10.64QNFCEE29 pKa = 4.14RR30 pKa = 11.84VAKK33 pKa = 9.38FAVMGRR39 pKa = 11.84EE40 pKa = 3.95IGSGGVPHH48 pKa = 6.74LQGFISLQNKK58 pKa = 8.17VRR60 pKa = 11.84LTGIKK65 pKa = 10.22AIVGQRR71 pKa = 11.84AHH73 pKa = 6.62VEE75 pKa = 3.97PAKK78 pKa = 10.45GTDD81 pKa = 3.37QKK83 pKa = 11.66NLEE86 pKa = 4.5YY87 pKa = 9.48CTKK90 pKa = 10.5SCGVDD95 pKa = 2.84GATIYY100 pKa = 10.07GVPAGGVTEE109 pKa = 4.57KK110 pKa = 11.23GGGTPLSRR118 pKa = 11.84RR119 pKa = 11.84ALEE122 pKa = 4.22CVRR125 pKa = 11.84KK126 pKa = 9.92RR127 pKa = 11.84SAGTSISEE135 pKa = 4.65LLDD138 pKa = 4.63DD139 pKa = 4.84DD140 pKa = 4.72TLAPAYY146 pKa = 9.14IVHH149 pKa = 6.28KK150 pKa = 10.49RR151 pKa = 11.84AIEE154 pKa = 3.99EE155 pKa = 4.31CASDD159 pKa = 3.45IAGAQQFQAEE169 pKa = 4.41RR170 pKa = 11.84ARR172 pKa = 11.84YY173 pKa = 7.45ATTRR177 pKa = 11.84WKK179 pKa = 10.24PFQFNILKK187 pKa = 9.78ICMQEE192 pKa = 3.51PHH194 pKa = 6.78PRR196 pKa = 11.84AVHH199 pKa = 5.65WFWDD203 pKa = 3.69AEE205 pKa = 4.35GNTGKK210 pKa = 8.27TFIAKK215 pKa = 10.05YY216 pKa = 10.3LVLLHH221 pKa = 6.8DD222 pKa = 4.37AVRR225 pKa = 11.84FEE227 pKa = 4.45NGKK230 pKa = 9.75SADD233 pKa = 2.76IKK235 pKa = 10.72YY236 pKa = 10.06AYY238 pKa = 9.1KK239 pKa = 10.15GQRR242 pKa = 11.84IVIFDD247 pKa = 3.87FSRR250 pKa = 11.84SQTDD254 pKa = 3.58HH255 pKa = 6.52INYY258 pKa = 9.98EE259 pKa = 3.95VMEE262 pKa = 4.34SVKK265 pKa = 10.85NGIVFSPKK273 pKa = 9.53YY274 pKa = 8.59EE275 pKa = 4.0SGMKK279 pKa = 9.92VFRR282 pKa = 11.84TPHH285 pKa = 6.14MICFANEE292 pKa = 3.92RR293 pKa = 11.84PDD295 pKa = 3.76TSNMSMDD302 pKa = 2.64RR303 pKa = 11.84WEE305 pKa = 3.73IHH307 pKa = 6.99EE308 pKa = 4.04ITTRR312 pKa = 11.84QARR315 pKa = 11.84TPTTLPDD322 pKa = 3.88RR323 pKa = 11.84DD324 pKa = 3.87SDD326 pKa = 3.79THH328 pKa = 6.7EE329 pKa = 4.01IAA331 pKa = 5.06
Molecular weight: 37.46 kDa
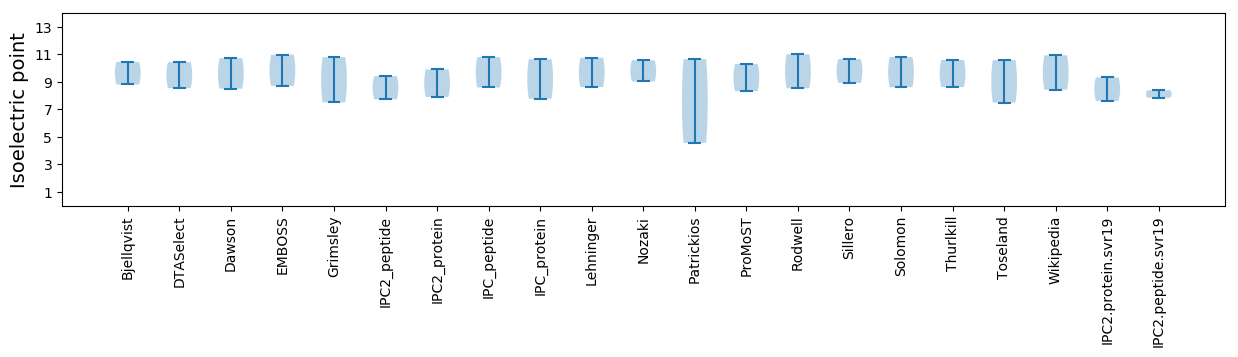
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L7UX06|L7UX06_9VIRU Putative capsid protein OS=Cyanoramphus nest associated circular X DNA virus OX=1282443 PE=4 SV=1
MM1 pKa = 6.94GRR3 pKa = 11.84KK4 pKa = 8.64RR5 pKa = 11.84AAPWAGKK12 pKa = 10.26SYY14 pKa = 9.92GKK16 pKa = 9.72RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84VIRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84WPVPARR31 pKa = 11.84KK32 pKa = 9.3RR33 pKa = 11.84RR34 pKa = 11.84VPKK37 pKa = 9.83RR38 pKa = 11.84RR39 pKa = 11.84SRR41 pKa = 11.84RR42 pKa = 11.84TRR44 pKa = 11.84KK45 pKa = 5.5TTRR48 pKa = 11.84DD49 pKa = 3.02NARR52 pKa = 11.84VYY54 pKa = 10.57RR55 pKa = 11.84QGLFARR61 pKa = 11.84FIGPTYY67 pKa = 9.8TLSAAANEE75 pKa = 4.24DD76 pKa = 3.08AVYY79 pKa = 9.76TLNFNYY85 pKa = 8.0GTLGQNDD92 pKa = 3.93FAKK95 pKa = 10.61QMEE98 pKa = 5.3GYY100 pKa = 11.16ARR102 pKa = 11.84MFEE105 pKa = 4.24QMKK108 pKa = 10.25CYY110 pKa = 9.64KK111 pKa = 10.66AKK113 pKa = 9.35LTYY116 pKa = 8.9WLNDD120 pKa = 3.08NDD122 pKa = 4.0EE123 pKa = 4.19LAKK126 pKa = 10.72PEE128 pKa = 4.37TKK130 pKa = 8.29WTEE133 pKa = 3.9VVTSYY138 pKa = 11.43DD139 pKa = 3.19PRR141 pKa = 11.84NYY143 pKa = 9.68ARR145 pKa = 11.84NMTFQGALSRR155 pKa = 11.84GNSKK159 pKa = 10.35RR160 pKa = 11.84VILKK164 pKa = 9.72PGRR167 pKa = 11.84KK168 pKa = 7.15YY169 pKa = 9.32TATITPKK176 pKa = 10.02WGVNLAAADD185 pKa = 3.95VPAPSDD191 pKa = 3.44PSVVQVGGRR200 pKa = 11.84SGYY203 pKa = 10.51VNMSRR208 pKa = 11.84VALATDD214 pKa = 3.48ISINSLCMVFHH225 pKa = 6.63GMPAEE230 pKa = 4.11TDD232 pKa = 3.95FLSCYY237 pKa = 8.45NTFYY241 pKa = 11.14CHH243 pKa = 6.92FKK245 pKa = 11.01DD246 pKa = 3.28RR247 pKa = 11.84VQMNPYY253 pKa = 10.04KK254 pKa = 10.74LL255 pKa = 3.56
MM1 pKa = 6.94GRR3 pKa = 11.84KK4 pKa = 8.64RR5 pKa = 11.84AAPWAGKK12 pKa = 10.26SYY14 pKa = 9.92GKK16 pKa = 9.72RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84VIRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84WPVPARR31 pKa = 11.84KK32 pKa = 9.3RR33 pKa = 11.84RR34 pKa = 11.84VPKK37 pKa = 9.83RR38 pKa = 11.84RR39 pKa = 11.84SRR41 pKa = 11.84RR42 pKa = 11.84TRR44 pKa = 11.84KK45 pKa = 5.5TTRR48 pKa = 11.84DD49 pKa = 3.02NARR52 pKa = 11.84VYY54 pKa = 10.57RR55 pKa = 11.84QGLFARR61 pKa = 11.84FIGPTYY67 pKa = 9.8TLSAAANEE75 pKa = 4.24DD76 pKa = 3.08AVYY79 pKa = 9.76TLNFNYY85 pKa = 8.0GTLGQNDD92 pKa = 3.93FAKK95 pKa = 10.61QMEE98 pKa = 5.3GYY100 pKa = 11.16ARR102 pKa = 11.84MFEE105 pKa = 4.24QMKK108 pKa = 10.25CYY110 pKa = 9.64KK111 pKa = 10.66AKK113 pKa = 9.35LTYY116 pKa = 8.9WLNDD120 pKa = 3.08NDD122 pKa = 4.0EE123 pKa = 4.19LAKK126 pKa = 10.72PEE128 pKa = 4.37TKK130 pKa = 8.29WTEE133 pKa = 3.9VVTSYY138 pKa = 11.43DD139 pKa = 3.19PRR141 pKa = 11.84NYY143 pKa = 9.68ARR145 pKa = 11.84NMTFQGALSRR155 pKa = 11.84GNSKK159 pKa = 10.35RR160 pKa = 11.84VILKK164 pKa = 9.72PGRR167 pKa = 11.84KK168 pKa = 7.15YY169 pKa = 9.32TATITPKK176 pKa = 10.02WGVNLAAADD185 pKa = 3.95VPAPSDD191 pKa = 3.44PSVVQVGGRR200 pKa = 11.84SGYY203 pKa = 10.51VNMSRR208 pKa = 11.84VALATDD214 pKa = 3.48ISINSLCMVFHH225 pKa = 6.63GMPAEE230 pKa = 4.11TDD232 pKa = 3.95FLSCYY237 pKa = 8.45NTFYY241 pKa = 11.14CHH243 pKa = 6.92FKK245 pKa = 11.01DD246 pKa = 3.28RR247 pKa = 11.84VQMNPYY253 pKa = 10.04KK254 pKa = 10.74LL255 pKa = 3.56
Molecular weight: 29.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
586 |
255 |
331 |
293.0 |
33.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.703 ± 0.41 | 2.048 ± 0.277 |
4.778 ± 0.268 | 4.949 ± 1.273 |
4.608 ± 0.396 | 6.826 ± 0.319 |
2.048 ± 0.73 | 4.778 ± 1.401 |
6.655 ± 0.233 | 4.778 ± 0.411 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.901 ± 0.363 | 4.949 ± 0.539 |
4.608 ± 0.51 | 3.584 ± 0.485 |
9.215 ± 1.473 | 5.29 ± 0.111 |
6.997 ± 0.036 | 6.314 ± 0.204 |
1.706 ± 0.147 | 4.266 ± 0.934 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
