
Alces alces faeces associated circular virus MP65
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; unclassified Genomoviridae
Average proteome isoelectric point is 7.71
Get precalculated fractions of proteins
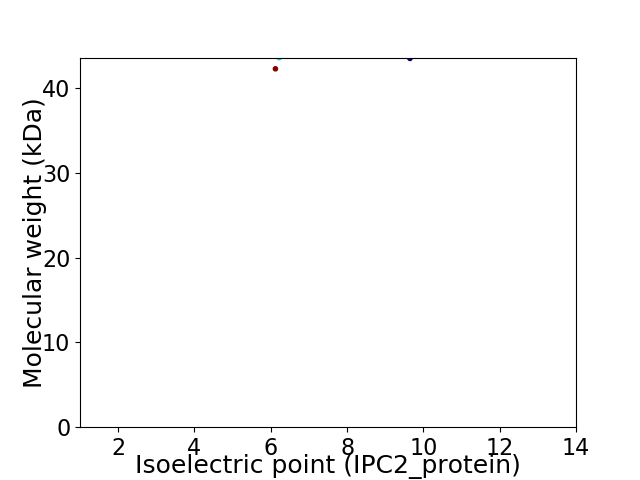
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
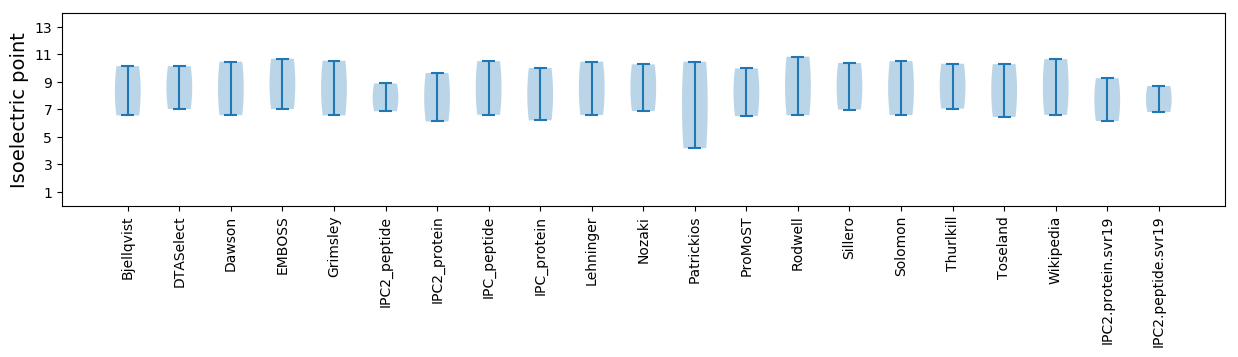
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5CK43|A0A2Z5CK43_9VIRU Replication associated protein OS=Alces alces faeces associated circular virus MP65 OX=2219143 PE=4 SV=1
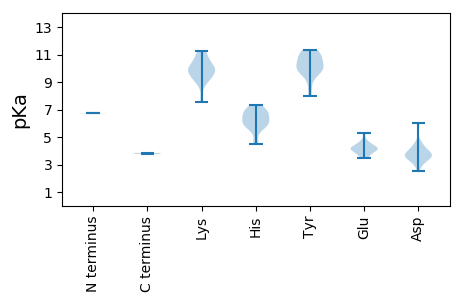
MM1 pKa = 6.73VQQIADD7 pKa = 3.8GLAAGAQEE15 pKa = 4.31PADD18 pKa = 3.61IPGVTGPGQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.42GTFRR34 pKa = 11.84CKK36 pKa = 9.31NQRR39 pKa = 11.84LFLTYY44 pKa = 10.41SQVGSGWDD52 pKa = 3.74PQSLTDD58 pKa = 3.94DD59 pKa = 4.32LEE61 pKa = 4.83RR62 pKa = 11.84LGAVHH67 pKa = 7.35RR68 pKa = 11.84LGKK71 pKa = 9.75EE72 pKa = 3.51LHH74 pKa = 5.9QDD76 pKa = 3.14GGIHH80 pKa = 4.51YY81 pKa = 9.68HH82 pKa = 6.77CYY84 pKa = 10.42VDD86 pKa = 4.54FGRR89 pKa = 11.84PLEE92 pKa = 4.33FEE94 pKa = 4.22NCHH97 pKa = 5.68KK98 pKa = 10.32FCYY101 pKa = 9.92GGPPSAGCPKK111 pKa = 10.39GGHH114 pKa = 6.08CNILVIRR121 pKa = 11.84RR122 pKa = 11.84TPHH125 pKa = 6.14HH126 pKa = 6.24AWDD129 pKa = 3.87YY130 pKa = 10.83AGKK133 pKa = 9.98DD134 pKa = 3.08GDD136 pKa = 4.57VISEE140 pKa = 4.0NCPRR144 pKa = 11.84PLVVKK149 pKa = 9.97GQSEE153 pKa = 4.58RR154 pKa = 11.84KK155 pKa = 9.96AEE157 pKa = 3.77WVYY160 pKa = 11.33CLDD163 pKa = 3.35ATGFDD168 pKa = 3.54NFYY171 pKa = 11.31EE172 pKa = 4.53RR173 pKa = 11.84IKK175 pKa = 11.15DD176 pKa = 3.49VDD178 pKa = 3.64PASLVKK184 pKa = 10.63SSFSVEE190 pKa = 3.72HH191 pKa = 5.79CAKK194 pKa = 10.4RR195 pKa = 11.84LFRR198 pKa = 11.84QRR200 pKa = 11.84QPEE203 pKa = 4.52TFTPIDD209 pKa = 3.48GLQFHH214 pKa = 6.85WEE216 pKa = 4.22SFPKK220 pKa = 9.06VAQWVLRR227 pKa = 11.84VLPDD231 pKa = 3.32DD232 pKa = 3.92RR233 pKa = 11.84QLVGGRR239 pKa = 11.84RR240 pKa = 11.84PSLILWGASLHH251 pKa = 6.3GKK253 pKa = 7.84TDD255 pKa = 4.02LARR258 pKa = 11.84SLGNHH263 pKa = 4.53IHH265 pKa = 6.72FGNDD269 pKa = 2.79FSLAEE274 pKa = 4.01LLAVGVEE281 pKa = 4.14NVDD284 pKa = 4.55FGVLDD289 pKa = 4.34DD290 pKa = 6.0LDD292 pKa = 3.73WTDD295 pKa = 5.92PILKK299 pKa = 9.48GNKK302 pKa = 7.52YY303 pKa = 9.94KK304 pKa = 10.86AWLGCQEE311 pKa = 5.27HH312 pKa = 6.83FICTDD317 pKa = 2.54KK318 pKa = 11.22YY319 pKa = 11.0LRR321 pKa = 11.84KK322 pKa = 9.9EE323 pKa = 4.04SIKK326 pKa = 9.87WGKK329 pKa = 8.99PCIFLSNDD337 pKa = 3.53DD338 pKa = 3.67PTEE341 pKa = 4.12SLKK344 pKa = 11.15HH345 pKa = 6.8SDD347 pKa = 4.04FLWIQKK353 pKa = 9.15NCWIVEE359 pKa = 4.26VPDD362 pKa = 3.77GQRR365 pKa = 11.84IASRR369 pKa = 11.84PSVFEE374 pKa = 3.85
MM1 pKa = 6.73VQQIADD7 pKa = 3.8GLAAGAQEE15 pKa = 4.31PADD18 pKa = 3.61IPGVTGPGQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.42GTFRR34 pKa = 11.84CKK36 pKa = 9.31NQRR39 pKa = 11.84LFLTYY44 pKa = 10.41SQVGSGWDD52 pKa = 3.74PQSLTDD58 pKa = 3.94DD59 pKa = 4.32LEE61 pKa = 4.83RR62 pKa = 11.84LGAVHH67 pKa = 7.35RR68 pKa = 11.84LGKK71 pKa = 9.75EE72 pKa = 3.51LHH74 pKa = 5.9QDD76 pKa = 3.14GGIHH80 pKa = 4.51YY81 pKa = 9.68HH82 pKa = 6.77CYY84 pKa = 10.42VDD86 pKa = 4.54FGRR89 pKa = 11.84PLEE92 pKa = 4.33FEE94 pKa = 4.22NCHH97 pKa = 5.68KK98 pKa = 10.32FCYY101 pKa = 9.92GGPPSAGCPKK111 pKa = 10.39GGHH114 pKa = 6.08CNILVIRR121 pKa = 11.84RR122 pKa = 11.84TPHH125 pKa = 6.14HH126 pKa = 6.24AWDD129 pKa = 3.87YY130 pKa = 10.83AGKK133 pKa = 9.98DD134 pKa = 3.08GDD136 pKa = 4.57VISEE140 pKa = 4.0NCPRR144 pKa = 11.84PLVVKK149 pKa = 9.97GQSEE153 pKa = 4.58RR154 pKa = 11.84KK155 pKa = 9.96AEE157 pKa = 3.77WVYY160 pKa = 11.33CLDD163 pKa = 3.35ATGFDD168 pKa = 3.54NFYY171 pKa = 11.31EE172 pKa = 4.53RR173 pKa = 11.84IKK175 pKa = 11.15DD176 pKa = 3.49VDD178 pKa = 3.64PASLVKK184 pKa = 10.63SSFSVEE190 pKa = 3.72HH191 pKa = 5.79CAKK194 pKa = 10.4RR195 pKa = 11.84LFRR198 pKa = 11.84QRR200 pKa = 11.84QPEE203 pKa = 4.52TFTPIDD209 pKa = 3.48GLQFHH214 pKa = 6.85WEE216 pKa = 4.22SFPKK220 pKa = 9.06VAQWVLRR227 pKa = 11.84VLPDD231 pKa = 3.32DD232 pKa = 3.92RR233 pKa = 11.84QLVGGRR239 pKa = 11.84RR240 pKa = 11.84PSLILWGASLHH251 pKa = 6.3GKK253 pKa = 7.84TDD255 pKa = 4.02LARR258 pKa = 11.84SLGNHH263 pKa = 4.53IHH265 pKa = 6.72FGNDD269 pKa = 2.79FSLAEE274 pKa = 4.01LLAVGVEE281 pKa = 4.14NVDD284 pKa = 4.55FGVLDD289 pKa = 4.34DD290 pKa = 6.0LDD292 pKa = 3.73WTDD295 pKa = 5.92PILKK299 pKa = 9.48GNKK302 pKa = 7.52YY303 pKa = 9.94KK304 pKa = 10.86AWLGCQEE311 pKa = 5.27HH312 pKa = 6.83FICTDD317 pKa = 2.54KK318 pKa = 11.22YY319 pKa = 11.0LRR321 pKa = 11.84KK322 pKa = 9.9EE323 pKa = 4.04SIKK326 pKa = 9.87WGKK329 pKa = 8.99PCIFLSNDD337 pKa = 3.53DD338 pKa = 3.67PTEE341 pKa = 4.12SLKK344 pKa = 11.15HH345 pKa = 6.8SDD347 pKa = 4.04FLWIQKK353 pKa = 9.15NCWIVEE359 pKa = 4.26VPDD362 pKa = 3.77GQRR365 pKa = 11.84IASRR369 pKa = 11.84PSVFEE374 pKa = 3.85
Molecular weight: 42.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CK43|A0A2Z5CK43_9VIRU Replication associated protein OS=Alces alces faeces associated circular virus MP65 OX=2219143 PE=4 SV=1
MM1 pKa = 6.75NQARR5 pKa = 11.84RR6 pKa = 11.84DD7 pKa = 3.55RR8 pKa = 11.84EE9 pKa = 3.69LALFQRR15 pKa = 11.84GPDD18 pKa = 3.13LWINRR23 pKa = 11.84LASHH27 pKa = 6.03TTRR30 pKa = 11.84TDD32 pKa = 3.11DD33 pKa = 3.78PLGIKK38 pKa = 10.41YY39 pKa = 9.96EE40 pKa = 4.15VGLPSTPRR48 pKa = 11.84EE49 pKa = 3.52ARR51 pKa = 11.84LFVFQIFGQHH61 pKa = 5.74RR62 pKa = 11.84RR63 pKa = 11.84TLPLVLKK70 pKa = 10.56GLEE73 pKa = 4.02RR74 pKa = 11.84EE75 pKa = 4.55LNGTAPFYY83 pKa = 10.79PRR85 pKa = 11.84MVSSQATALAQLWKK99 pKa = 9.87NKK101 pKa = 9.28KK102 pKa = 8.67MPAKK106 pKa = 9.71KK107 pKa = 9.9SSRR110 pKa = 11.84GGIGKK115 pKa = 9.13AAKK118 pKa = 9.82AKK120 pKa = 9.95KK121 pKa = 10.34NYY123 pKa = 9.48GATPAFNRR131 pKa = 11.84ALTAAVQKK139 pKa = 10.98VEE141 pKa = 4.2ARR143 pKa = 11.84SQEE146 pKa = 4.02TTYY149 pKa = 10.74SQSGVYY155 pKa = 10.25LNAAQSTFTQTGYY168 pKa = 8.74TLSGLNQSAAYY179 pKa = 9.87AVGTTNTRR187 pKa = 11.84ANATNQVFAFPICPLAQVGNSSTPGYY213 pKa = 10.36RR214 pKa = 11.84KK215 pKa = 9.34GQRR218 pKa = 11.84INPVGFRR225 pKa = 11.84FSIQHH230 pKa = 5.85DD231 pKa = 3.63QSLATMSATYY241 pKa = 9.86KK242 pKa = 9.42WALIRR247 pKa = 11.84NQGGTLSGNTGPVITQTNAITMFVGLVQGPLASAGGPNGALPNGDD292 pKa = 3.99FASSMRR298 pKa = 11.84WNRR301 pKa = 11.84QAWTVKK307 pKa = 10.2KK308 pKa = 10.07QGSYY312 pKa = 9.84TMPAALARR320 pKa = 11.84DD321 pKa = 4.13NNTAPASATAPPFHH335 pKa = 6.95NSSKK339 pKa = 9.63TVTHH343 pKa = 7.11FIPIRR348 pKa = 11.84DD349 pKa = 3.95SHH351 pKa = 6.55WDD353 pKa = 3.61YY354 pKa = 7.96PTPTAIANIKK364 pKa = 9.76GGDD367 pKa = 3.61YY368 pKa = 11.13YY369 pKa = 11.08FVLWRR374 pKa = 11.84EE375 pKa = 3.82GSADD379 pKa = 3.49QFIGNDD385 pKa = 4.43FINFICEE392 pKa = 4.25LSFKK396 pKa = 11.03DD397 pKa = 3.55PP398 pKa = 3.77
MM1 pKa = 6.75NQARR5 pKa = 11.84RR6 pKa = 11.84DD7 pKa = 3.55RR8 pKa = 11.84EE9 pKa = 3.69LALFQRR15 pKa = 11.84GPDD18 pKa = 3.13LWINRR23 pKa = 11.84LASHH27 pKa = 6.03TTRR30 pKa = 11.84TDD32 pKa = 3.11DD33 pKa = 3.78PLGIKK38 pKa = 10.41YY39 pKa = 9.96EE40 pKa = 4.15VGLPSTPRR48 pKa = 11.84EE49 pKa = 3.52ARR51 pKa = 11.84LFVFQIFGQHH61 pKa = 5.74RR62 pKa = 11.84RR63 pKa = 11.84TLPLVLKK70 pKa = 10.56GLEE73 pKa = 4.02RR74 pKa = 11.84EE75 pKa = 4.55LNGTAPFYY83 pKa = 10.79PRR85 pKa = 11.84MVSSQATALAQLWKK99 pKa = 9.87NKK101 pKa = 9.28KK102 pKa = 8.67MPAKK106 pKa = 9.71KK107 pKa = 9.9SSRR110 pKa = 11.84GGIGKK115 pKa = 9.13AAKK118 pKa = 9.82AKK120 pKa = 9.95KK121 pKa = 10.34NYY123 pKa = 9.48GATPAFNRR131 pKa = 11.84ALTAAVQKK139 pKa = 10.98VEE141 pKa = 4.2ARR143 pKa = 11.84SQEE146 pKa = 4.02TTYY149 pKa = 10.74SQSGVYY155 pKa = 10.25LNAAQSTFTQTGYY168 pKa = 8.74TLSGLNQSAAYY179 pKa = 9.87AVGTTNTRR187 pKa = 11.84ANATNQVFAFPICPLAQVGNSSTPGYY213 pKa = 10.36RR214 pKa = 11.84KK215 pKa = 9.34GQRR218 pKa = 11.84INPVGFRR225 pKa = 11.84FSIQHH230 pKa = 5.85DD231 pKa = 3.63QSLATMSATYY241 pKa = 9.86KK242 pKa = 9.42WALIRR247 pKa = 11.84NQGGTLSGNTGPVITQTNAITMFVGLVQGPLASAGGPNGALPNGDD292 pKa = 3.99FASSMRR298 pKa = 11.84WNRR301 pKa = 11.84QAWTVKK307 pKa = 10.2KK308 pKa = 10.07QGSYY312 pKa = 9.84TMPAALARR320 pKa = 11.84DD321 pKa = 4.13NNTAPASATAPPFHH335 pKa = 6.95NSSKK339 pKa = 9.63TVTHH343 pKa = 7.11FIPIRR348 pKa = 11.84DD349 pKa = 3.95SHH351 pKa = 6.55WDD353 pKa = 3.61YY354 pKa = 7.96PTPTAIANIKK364 pKa = 9.76GGDD367 pKa = 3.61YY368 pKa = 11.13YY369 pKa = 11.08FVLWRR374 pKa = 11.84EE375 pKa = 3.82GSADD379 pKa = 3.49QFIGNDD385 pKa = 4.43FINFICEE392 pKa = 4.25LSFKK396 pKa = 11.03DD397 pKa = 3.55PP398 pKa = 3.77
Molecular weight: 43.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
772 |
374 |
398 |
386.0 |
42.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.549 ± 2.24 | 1.943 ± 1.072 |
5.44 ± 1.618 | 3.627 ± 1.017 |
5.052 ± 0.02 | 8.808 ± 0.385 |
2.72 ± 0.903 | 4.404 ± 0.099 |
5.311 ± 0.4 | 8.031 ± 0.741 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.036 ± 0.538 | 4.663 ± 1.205 |
6.088 ± 0.144 | 5.181 ± 0.445 |
6.218 ± 0.047 | 6.347 ± 0.512 |
6.088 ± 2.014 | 5.311 ± 0.774 |
2.332 ± 0.426 | 2.85 ± 0.31 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
