
Alistipes dispar
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
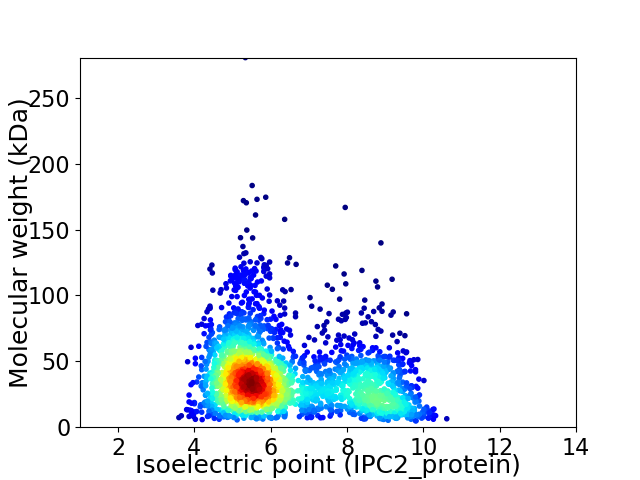
Virtual 2D-PAGE plot for 2408 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y1X0X4|A0A4Y1X0X4_9BACT Nudix hydrolase domain-containing protein OS=Alistipes dispar OX=2585119 GN=A5CPEGH6_05960 PE=4 SV=1
MM1 pKa = 8.02ALMLALPLFAAGCIIDD17 pKa = 4.88DD18 pKa = 4.58PEE20 pKa = 4.96PGPNLSGDD28 pKa = 3.06WTEE31 pKa = 4.33PEE33 pKa = 4.3EE34 pKa = 4.21PAGPEE39 pKa = 3.68TFITVDD45 pKa = 3.59GDD47 pKa = 3.61FSDD50 pKa = 3.99WDD52 pKa = 4.63AIPADD57 pKa = 4.16EE58 pKa = 5.21LVTATVPDD66 pKa = 3.99DD67 pKa = 3.62AFYY70 pKa = 11.16SAGRR74 pKa = 11.84RR75 pKa = 11.84LKK77 pKa = 10.15VYY79 pKa = 10.9AGATYY84 pKa = 10.37INIYY88 pKa = 10.3LEE90 pKa = 4.18YY91 pKa = 10.67SEE93 pKa = 6.13AEE95 pKa = 3.89QPVQIMHH102 pKa = 7.34LYY104 pKa = 10.05LDD106 pKa = 3.6SDD108 pKa = 3.8MAFDD112 pKa = 4.28GEE114 pKa = 4.43GRR116 pKa = 11.84PTTGMGNAAWTNDD129 pKa = 3.1GADD132 pKa = 3.2ILFEE136 pKa = 4.31GMLRR140 pKa = 11.84DD141 pKa = 3.86DD142 pKa = 4.04TGAPTAYY149 pKa = 10.26APTIFTWSGEE159 pKa = 4.05PLSGGWDD166 pKa = 3.07WTEE169 pKa = 3.54AVAAGAGICSNCEE182 pKa = 3.68PVEE185 pKa = 4.01LPNGNMAIEE194 pKa = 4.09MTIARR199 pKa = 11.84SAVPGLGQSFRR210 pKa = 11.84LGVGLQYY217 pKa = 11.21DD218 pKa = 4.01WNDD221 pKa = 2.95IAYY224 pKa = 9.3LPAGSAVEE232 pKa = 4.11EE233 pKa = 4.2NGEE236 pKa = 4.15PKK238 pKa = 10.45HH239 pKa = 6.05GAVEE243 pKa = 3.96NLLIGAKK250 pKa = 9.8SGPAPEE256 pKa = 4.23VLITADD262 pKa = 3.88GDD264 pKa = 3.67FSDD267 pKa = 4.15WEE269 pKa = 4.26KK270 pKa = 11.24VPASEE275 pKa = 4.2MTVATVPDD283 pKa = 3.73DD284 pKa = 4.08AYY286 pKa = 11.38YY287 pKa = 10.94KK288 pKa = 10.07AGKK291 pKa = 8.12VAKK294 pKa = 9.53VYY296 pKa = 10.86AGEE299 pKa = 4.41TYY301 pKa = 10.06VNFYY305 pKa = 11.11VEE307 pKa = 4.63YY308 pKa = 9.51EE309 pKa = 4.28DD310 pKa = 5.45SAEE313 pKa = 3.95QPVQIMHH320 pKa = 7.34LYY322 pKa = 10.05LDD324 pKa = 3.6SDD326 pKa = 3.8MAFDD330 pKa = 4.28GEE332 pKa = 4.43GRR334 pKa = 11.84PTTGMGNAAWTNDD347 pKa = 3.1GADD350 pKa = 3.16ILFEE354 pKa = 4.16GMVVDD359 pKa = 5.62DD360 pKa = 5.21AGAPAAYY367 pKa = 10.01DD368 pKa = 3.32PTIFNWTGEE377 pKa = 4.2NLSGGWDD384 pKa = 3.31WTEE387 pKa = 3.66LLAAGSGVCSASKK400 pKa = 10.14PVEE403 pKa = 3.93LEE405 pKa = 3.72NGRR408 pKa = 11.84KK409 pKa = 8.86AVEE412 pKa = 4.2VVVVRR417 pKa = 11.84SSIPNLGSRR426 pKa = 11.84FRR428 pKa = 11.84VGIGLQYY435 pKa = 11.01DD436 pKa = 3.83WNDD439 pKa = 2.76IAYY442 pKa = 8.67IPAGSAVEE450 pKa = 3.99EE451 pKa = 4.31NGVVGHH457 pKa = 6.15GAVEE461 pKa = 3.94NMLVGRR467 pKa = 4.74
MM1 pKa = 8.02ALMLALPLFAAGCIIDD17 pKa = 4.88DD18 pKa = 4.58PEE20 pKa = 4.96PGPNLSGDD28 pKa = 3.06WTEE31 pKa = 4.33PEE33 pKa = 4.3EE34 pKa = 4.21PAGPEE39 pKa = 3.68TFITVDD45 pKa = 3.59GDD47 pKa = 3.61FSDD50 pKa = 3.99WDD52 pKa = 4.63AIPADD57 pKa = 4.16EE58 pKa = 5.21LVTATVPDD66 pKa = 3.99DD67 pKa = 3.62AFYY70 pKa = 11.16SAGRR74 pKa = 11.84RR75 pKa = 11.84LKK77 pKa = 10.15VYY79 pKa = 10.9AGATYY84 pKa = 10.37INIYY88 pKa = 10.3LEE90 pKa = 4.18YY91 pKa = 10.67SEE93 pKa = 6.13AEE95 pKa = 3.89QPVQIMHH102 pKa = 7.34LYY104 pKa = 10.05LDD106 pKa = 3.6SDD108 pKa = 3.8MAFDD112 pKa = 4.28GEE114 pKa = 4.43GRR116 pKa = 11.84PTTGMGNAAWTNDD129 pKa = 3.1GADD132 pKa = 3.2ILFEE136 pKa = 4.31GMLRR140 pKa = 11.84DD141 pKa = 3.86DD142 pKa = 4.04TGAPTAYY149 pKa = 10.26APTIFTWSGEE159 pKa = 4.05PLSGGWDD166 pKa = 3.07WTEE169 pKa = 3.54AVAAGAGICSNCEE182 pKa = 3.68PVEE185 pKa = 4.01LPNGNMAIEE194 pKa = 4.09MTIARR199 pKa = 11.84SAVPGLGQSFRR210 pKa = 11.84LGVGLQYY217 pKa = 11.21DD218 pKa = 4.01WNDD221 pKa = 2.95IAYY224 pKa = 9.3LPAGSAVEE232 pKa = 4.11EE233 pKa = 4.2NGEE236 pKa = 4.15PKK238 pKa = 10.45HH239 pKa = 6.05GAVEE243 pKa = 3.96NLLIGAKK250 pKa = 9.8SGPAPEE256 pKa = 4.23VLITADD262 pKa = 3.88GDD264 pKa = 3.67FSDD267 pKa = 4.15WEE269 pKa = 4.26KK270 pKa = 11.24VPASEE275 pKa = 4.2MTVATVPDD283 pKa = 3.73DD284 pKa = 4.08AYY286 pKa = 11.38YY287 pKa = 10.94KK288 pKa = 10.07AGKK291 pKa = 8.12VAKK294 pKa = 9.53VYY296 pKa = 10.86AGEE299 pKa = 4.41TYY301 pKa = 10.06VNFYY305 pKa = 11.11VEE307 pKa = 4.63YY308 pKa = 9.51EE309 pKa = 4.28DD310 pKa = 5.45SAEE313 pKa = 3.95QPVQIMHH320 pKa = 7.34LYY322 pKa = 10.05LDD324 pKa = 3.6SDD326 pKa = 3.8MAFDD330 pKa = 4.28GEE332 pKa = 4.43GRR334 pKa = 11.84PTTGMGNAAWTNDD347 pKa = 3.1GADD350 pKa = 3.16ILFEE354 pKa = 4.16GMVVDD359 pKa = 5.62DD360 pKa = 5.21AGAPAAYY367 pKa = 10.01DD368 pKa = 3.32PTIFNWTGEE377 pKa = 4.2NLSGGWDD384 pKa = 3.31WTEE387 pKa = 3.66LLAAGSGVCSASKK400 pKa = 10.14PVEE403 pKa = 3.93LEE405 pKa = 3.72NGRR408 pKa = 11.84KK409 pKa = 8.86AVEE412 pKa = 4.2VVVVRR417 pKa = 11.84SSIPNLGSRR426 pKa = 11.84FRR428 pKa = 11.84VGIGLQYY435 pKa = 11.01DD436 pKa = 3.83WNDD439 pKa = 2.76IAYY442 pKa = 8.67IPAGSAVEE450 pKa = 3.99EE451 pKa = 4.31NGVVGHH457 pKa = 6.15GAVEE461 pKa = 3.94NMLVGRR467 pKa = 4.74
Molecular weight: 49.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y1X0M4|A0A4Y1X0M4_9BACT Uncharacterized protein OS=Alistipes dispar OX=2585119 GN=A5CPEGH6_04860 PE=4 SV=1
MM1 pKa = 7.32TARR4 pKa = 11.84CGGVGRR10 pKa = 11.84TLRR13 pKa = 11.84RR14 pKa = 11.84AAARR18 pKa = 11.84LLLVSLAALAAAACGRR34 pKa = 11.84TEE36 pKa = 3.92PEE38 pKa = 3.76AVRR41 pKa = 11.84VEE43 pKa = 3.81AVEE46 pKa = 3.94RR47 pKa = 11.84FEE49 pKa = 4.78RR50 pKa = 11.84LGSAGADD57 pKa = 2.62AVLRR61 pKa = 11.84VHH63 pKa = 6.69NGTGHH68 pKa = 5.0TLKK71 pKa = 10.2IRR73 pKa = 11.84RR74 pKa = 11.84VRR76 pKa = 11.84LDD78 pKa = 2.83VFYY81 pKa = 10.86AGKK84 pKa = 10.33RR85 pKa = 11.84VGTLRR90 pKa = 11.84LHH92 pKa = 5.93EE93 pKa = 4.23PVRR96 pKa = 11.84IPRR99 pKa = 11.84RR100 pKa = 11.84ATTLVGTRR108 pKa = 11.84WRR110 pKa = 11.84LEE112 pKa = 3.93TSDD115 pKa = 3.73PMAFYY120 pKa = 10.72VVEE123 pKa = 4.08RR124 pKa = 11.84RR125 pKa = 11.84LGEE128 pKa = 4.2GDD130 pKa = 3.7LSRR133 pKa = 11.84IGLSYY138 pKa = 10.41EE139 pKa = 3.48LRR141 pKa = 11.84ARR143 pKa = 11.84SGVVPVKK150 pKa = 10.44ISGDD154 pKa = 3.56MMPLSEE160 pKa = 4.67FLNTFGLSEE169 pKa = 4.77DD170 pKa = 4.97DD171 pKa = 3.03IKK173 pKa = 11.4EE174 pKa = 4.01YY175 pKa = 11.2LKK177 pKa = 11.14
MM1 pKa = 7.32TARR4 pKa = 11.84CGGVGRR10 pKa = 11.84TLRR13 pKa = 11.84RR14 pKa = 11.84AAARR18 pKa = 11.84LLLVSLAALAAAACGRR34 pKa = 11.84TEE36 pKa = 3.92PEE38 pKa = 3.76AVRR41 pKa = 11.84VEE43 pKa = 3.81AVEE46 pKa = 3.94RR47 pKa = 11.84FEE49 pKa = 4.78RR50 pKa = 11.84LGSAGADD57 pKa = 2.62AVLRR61 pKa = 11.84VHH63 pKa = 6.69NGTGHH68 pKa = 5.0TLKK71 pKa = 10.2IRR73 pKa = 11.84RR74 pKa = 11.84VRR76 pKa = 11.84LDD78 pKa = 2.83VFYY81 pKa = 10.86AGKK84 pKa = 10.33RR85 pKa = 11.84VGTLRR90 pKa = 11.84LHH92 pKa = 5.93EE93 pKa = 4.23PVRR96 pKa = 11.84IPRR99 pKa = 11.84RR100 pKa = 11.84ATTLVGTRR108 pKa = 11.84WRR110 pKa = 11.84LEE112 pKa = 3.93TSDD115 pKa = 3.73PMAFYY120 pKa = 10.72VVEE123 pKa = 4.08RR124 pKa = 11.84RR125 pKa = 11.84LGEE128 pKa = 4.2GDD130 pKa = 3.7LSRR133 pKa = 11.84IGLSYY138 pKa = 10.41EE139 pKa = 3.48LRR141 pKa = 11.84ARR143 pKa = 11.84SGVVPVKK150 pKa = 10.44ISGDD154 pKa = 3.56MMPLSEE160 pKa = 4.67FLNTFGLSEE169 pKa = 4.77DD170 pKa = 4.97DD171 pKa = 3.03IKK173 pKa = 11.4EE174 pKa = 4.01YY175 pKa = 11.2LKK177 pKa = 11.14
Molecular weight: 19.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
882180 |
41 |
2498 |
366.4 |
40.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.709 ± 0.065 | 1.274 ± 0.018 |
5.782 ± 0.033 | 6.795 ± 0.047 |
4.231 ± 0.03 | 8.017 ± 0.039 |
1.71 ± 0.02 | 5.33 ± 0.045 |
4.119 ± 0.05 | 9.063 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.452 ± 0.023 | 3.566 ± 0.041 |
4.343 ± 0.033 | 2.735 ± 0.026 |
7.316 ± 0.064 | 5.584 ± 0.041 |
5.597 ± 0.041 | 7.066 ± 0.042 |
1.328 ± 0.023 | 3.982 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
