
Hubei toti-like virus 12
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
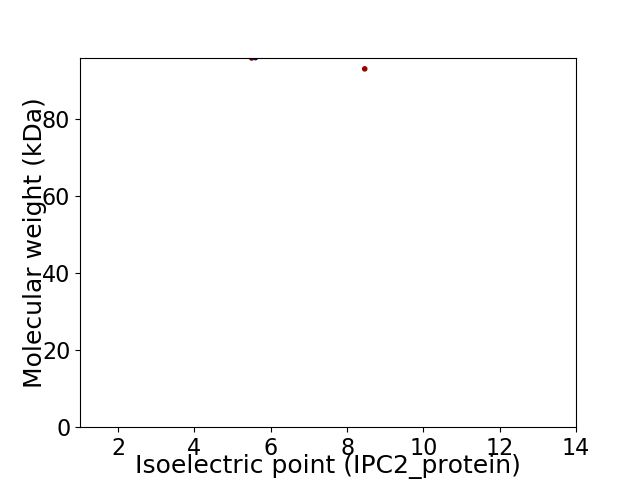
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KF57|A0A1L3KF57_9VIRU RdRp catalytic domain-containing protein OS=Hubei toti-like virus 12 OX=1923300 PE=4 SV=1
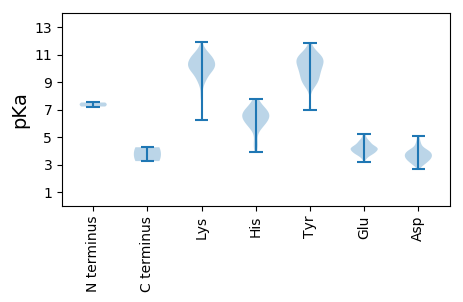
MM1 pKa = 7.51PADD4 pKa = 4.05LRR6 pKa = 11.84DD7 pKa = 3.78YY8 pKa = 10.06MCNCSSVAQTTGAHH22 pKa = 6.45ASLSLHH28 pKa = 6.28HH29 pKa = 6.57SVSRR33 pKa = 11.84LQVFAIDD40 pKa = 3.79VPGGEE45 pKa = 4.26VGIEE49 pKa = 3.76GGVNYY54 pKa = 10.6SIMRR58 pKa = 11.84RR59 pKa = 11.84IMPSTGLGCHH69 pKa = 5.81QGSPWVTSTFNEE81 pKa = 4.21VLQKK85 pKa = 10.62LVAPLEE91 pKa = 4.5STSGNYY97 pKa = 8.87GALVSNPAYY106 pKa = 10.85SDD108 pKa = 3.88FGSLAYY114 pKa = 9.41LTDD117 pKa = 3.64VLAKK121 pKa = 9.84STGSWLDD128 pKa = 3.04EE129 pKa = 4.35SPIVRR134 pKa = 11.84CLLLGDD140 pKa = 4.59LSVSHH145 pKa = 6.5RR146 pKa = 11.84TLQCDD151 pKa = 3.44PLIGQIVSDD160 pKa = 3.96GQIKK164 pKa = 9.93RR165 pKa = 11.84VEE167 pKa = 4.37RR168 pKa = 11.84YY169 pKa = 9.08KK170 pKa = 10.0PTYY173 pKa = 9.09PRR175 pKa = 11.84PDD177 pKa = 3.65DD178 pKa = 4.11ALNPAEE184 pKa = 4.19VQAMAVAMDD193 pKa = 3.74SWVMLRR199 pKa = 11.84TGQRR203 pKa = 11.84NVPDD207 pKa = 3.91WNVHH211 pKa = 4.78DD212 pKa = 4.12WSYY215 pKa = 11.86SVAIVPCEE223 pKa = 3.62TSMLEE228 pKa = 4.39SNSIWLYY235 pKa = 8.53MASFLTTEE243 pKa = 3.86IWNNRR248 pKa = 11.84YY249 pKa = 7.0VWKK252 pKa = 10.66VHH254 pKa = 4.48VQPRR258 pKa = 11.84SDD260 pKa = 3.26MSSEE264 pKa = 4.18EE265 pKa = 4.2PEE267 pKa = 4.04HH268 pKa = 7.34EE269 pKa = 4.26YY270 pKa = 10.77MLMVRR275 pKa = 11.84PRR277 pKa = 11.84AAQVEE282 pKa = 4.57VQGPGRR288 pKa = 11.84VMLVVTNITSRR299 pKa = 11.84SNLRR303 pKa = 11.84WVTIDD308 pKa = 3.67GFNIPVWNPASGIGPQSVDD327 pKa = 3.08IMPMLLQYY335 pKa = 11.27LDD337 pKa = 3.92LQGATNNISQNWLQAWKK354 pKa = 9.54WMCDD358 pKa = 2.99RR359 pKa = 11.84VSSGNALNHH368 pKa = 6.32ALFLAAEE375 pKa = 4.66TSRR378 pKa = 11.84VQHH381 pKa = 6.49IAPLAHH387 pKa = 6.52PAVEE391 pKa = 4.0NSMPFYY397 pKa = 10.71SINLRR402 pKa = 11.84KK403 pKa = 9.4EE404 pKa = 3.76LPRR407 pKa = 11.84ILKK410 pKa = 10.21SNWEE414 pKa = 3.86MRR416 pKa = 11.84NWADD420 pKa = 4.29VDD422 pKa = 3.79PDD424 pKa = 3.82PEE426 pKa = 4.27KK427 pKa = 10.79EE428 pKa = 3.8KK429 pKa = 11.19SRR431 pKa = 11.84IHH433 pKa = 6.93DD434 pKa = 4.11WIEE437 pKa = 3.39QSIAAWISPLCIHH450 pKa = 7.15SNLTCSFEE458 pKa = 4.43TAEE461 pKa = 4.23SPDD464 pKa = 3.27WSKK467 pKa = 11.49LDD469 pKa = 3.49WMRR472 pKa = 11.84HH473 pKa = 3.89KK474 pKa = 10.45HH475 pKa = 6.0GADD478 pKa = 3.1QTATCHH484 pKa = 5.93EE485 pKa = 4.47ATPDD489 pKa = 3.25LRR491 pKa = 11.84VARR494 pKa = 11.84FVGLIAVPTPIVYY507 pKa = 9.56IDD509 pKa = 3.58PVAVQEE515 pKa = 4.42SVSAIAGMQVAGCAWMQMVTGISWLRR541 pKa = 11.84WVLYY545 pKa = 8.93TAAQNDD551 pKa = 3.96ATRR554 pKa = 11.84MVIQDD559 pKa = 3.11RR560 pKa = 11.84MMNEE564 pKa = 3.56ATAHH568 pKa = 5.87HH569 pKa = 6.7VSSGRR574 pKa = 11.84PDD576 pKa = 3.32KK577 pKa = 10.95TWINLLDD584 pKa = 3.77KK585 pKa = 11.25VLGVQVKK592 pKa = 10.04FDD594 pKa = 4.07FPQLLKK600 pKa = 9.68NTPSVLPAWWQVKK613 pKa = 9.77FILQKK618 pKa = 10.17MNYY621 pKa = 9.04LMPGEE626 pKa = 4.15LEE628 pKa = 3.85RR629 pKa = 11.84RR630 pKa = 11.84QRR632 pKa = 11.84PVFKK636 pKa = 10.78LGVLEE641 pKa = 4.68SSMKK645 pKa = 10.39VGFYY649 pKa = 11.07ANDD652 pKa = 3.56SKK654 pKa = 11.01HH655 pKa = 4.33WQAVARR661 pKa = 11.84YY662 pKa = 7.13TSMLGVSEE670 pKa = 4.5TNDD673 pKa = 3.25VPLYY677 pKa = 10.23AANNEE682 pKa = 4.32MFSHH686 pKa = 6.7LTQSYY691 pKa = 9.58LQLNLTRR698 pKa = 11.84LFTMCAEE705 pKa = 4.34SVYY708 pKa = 10.42HH709 pKa = 7.28SNGTYY714 pKa = 10.09YY715 pKa = 11.09NPLTVVDD722 pKa = 4.39TACAITPVANSLHH735 pKa = 5.61TSIPAEE741 pKa = 3.8LLIIKK746 pKa = 7.91TKK748 pKa = 10.79AGAGTNVGDD757 pKa = 3.73QTVLGEE763 pKa = 4.68WPDD766 pKa = 3.86PWWDD770 pKa = 3.04WLLAGVRR777 pKa = 11.84AAIPFLGSSIPTAALVGAASGLHH800 pKa = 5.88SFLAEE805 pKa = 3.84KK806 pKa = 10.54LNPSRR811 pKa = 11.84AKK813 pKa = 9.81EE814 pKa = 3.85EE815 pKa = 3.86QMIARR820 pKa = 11.84AQATKK825 pKa = 10.56ALEE828 pKa = 4.16SSAEE832 pKa = 3.94AVHH835 pKa = 7.09DD836 pKa = 3.84VLSTGRR842 pKa = 11.84EE843 pKa = 3.72AVGRR847 pKa = 11.84ISKK850 pKa = 10.31AINDD854 pKa = 3.9FTSDD858 pKa = 3.54QLDD861 pKa = 3.68EE862 pKa = 4.26
MM1 pKa = 7.51PADD4 pKa = 4.05LRR6 pKa = 11.84DD7 pKa = 3.78YY8 pKa = 10.06MCNCSSVAQTTGAHH22 pKa = 6.45ASLSLHH28 pKa = 6.28HH29 pKa = 6.57SVSRR33 pKa = 11.84LQVFAIDD40 pKa = 3.79VPGGEE45 pKa = 4.26VGIEE49 pKa = 3.76GGVNYY54 pKa = 10.6SIMRR58 pKa = 11.84RR59 pKa = 11.84IMPSTGLGCHH69 pKa = 5.81QGSPWVTSTFNEE81 pKa = 4.21VLQKK85 pKa = 10.62LVAPLEE91 pKa = 4.5STSGNYY97 pKa = 8.87GALVSNPAYY106 pKa = 10.85SDD108 pKa = 3.88FGSLAYY114 pKa = 9.41LTDD117 pKa = 3.64VLAKK121 pKa = 9.84STGSWLDD128 pKa = 3.04EE129 pKa = 4.35SPIVRR134 pKa = 11.84CLLLGDD140 pKa = 4.59LSVSHH145 pKa = 6.5RR146 pKa = 11.84TLQCDD151 pKa = 3.44PLIGQIVSDD160 pKa = 3.96GQIKK164 pKa = 9.93RR165 pKa = 11.84VEE167 pKa = 4.37RR168 pKa = 11.84YY169 pKa = 9.08KK170 pKa = 10.0PTYY173 pKa = 9.09PRR175 pKa = 11.84PDD177 pKa = 3.65DD178 pKa = 4.11ALNPAEE184 pKa = 4.19VQAMAVAMDD193 pKa = 3.74SWVMLRR199 pKa = 11.84TGQRR203 pKa = 11.84NVPDD207 pKa = 3.91WNVHH211 pKa = 4.78DD212 pKa = 4.12WSYY215 pKa = 11.86SVAIVPCEE223 pKa = 3.62TSMLEE228 pKa = 4.39SNSIWLYY235 pKa = 8.53MASFLTTEE243 pKa = 3.86IWNNRR248 pKa = 11.84YY249 pKa = 7.0VWKK252 pKa = 10.66VHH254 pKa = 4.48VQPRR258 pKa = 11.84SDD260 pKa = 3.26MSSEE264 pKa = 4.18EE265 pKa = 4.2PEE267 pKa = 4.04HH268 pKa = 7.34EE269 pKa = 4.26YY270 pKa = 10.77MLMVRR275 pKa = 11.84PRR277 pKa = 11.84AAQVEE282 pKa = 4.57VQGPGRR288 pKa = 11.84VMLVVTNITSRR299 pKa = 11.84SNLRR303 pKa = 11.84WVTIDD308 pKa = 3.67GFNIPVWNPASGIGPQSVDD327 pKa = 3.08IMPMLLQYY335 pKa = 11.27LDD337 pKa = 3.92LQGATNNISQNWLQAWKK354 pKa = 9.54WMCDD358 pKa = 2.99RR359 pKa = 11.84VSSGNALNHH368 pKa = 6.32ALFLAAEE375 pKa = 4.66TSRR378 pKa = 11.84VQHH381 pKa = 6.49IAPLAHH387 pKa = 6.52PAVEE391 pKa = 4.0NSMPFYY397 pKa = 10.71SINLRR402 pKa = 11.84KK403 pKa = 9.4EE404 pKa = 3.76LPRR407 pKa = 11.84ILKK410 pKa = 10.21SNWEE414 pKa = 3.86MRR416 pKa = 11.84NWADD420 pKa = 4.29VDD422 pKa = 3.79PDD424 pKa = 3.82PEE426 pKa = 4.27KK427 pKa = 10.79EE428 pKa = 3.8KK429 pKa = 11.19SRR431 pKa = 11.84IHH433 pKa = 6.93DD434 pKa = 4.11WIEE437 pKa = 3.39QSIAAWISPLCIHH450 pKa = 7.15SNLTCSFEE458 pKa = 4.43TAEE461 pKa = 4.23SPDD464 pKa = 3.27WSKK467 pKa = 11.49LDD469 pKa = 3.49WMRR472 pKa = 11.84HH473 pKa = 3.89KK474 pKa = 10.45HH475 pKa = 6.0GADD478 pKa = 3.1QTATCHH484 pKa = 5.93EE485 pKa = 4.47ATPDD489 pKa = 3.25LRR491 pKa = 11.84VARR494 pKa = 11.84FVGLIAVPTPIVYY507 pKa = 9.56IDD509 pKa = 3.58PVAVQEE515 pKa = 4.42SVSAIAGMQVAGCAWMQMVTGISWLRR541 pKa = 11.84WVLYY545 pKa = 8.93TAAQNDD551 pKa = 3.96ATRR554 pKa = 11.84MVIQDD559 pKa = 3.11RR560 pKa = 11.84MMNEE564 pKa = 3.56ATAHH568 pKa = 5.87HH569 pKa = 6.7VSSGRR574 pKa = 11.84PDD576 pKa = 3.32KK577 pKa = 10.95TWINLLDD584 pKa = 3.77KK585 pKa = 11.25VLGVQVKK592 pKa = 10.04FDD594 pKa = 4.07FPQLLKK600 pKa = 9.68NTPSVLPAWWQVKK613 pKa = 9.77FILQKK618 pKa = 10.17MNYY621 pKa = 9.04LMPGEE626 pKa = 4.15LEE628 pKa = 3.85RR629 pKa = 11.84RR630 pKa = 11.84QRR632 pKa = 11.84PVFKK636 pKa = 10.78LGVLEE641 pKa = 4.68SSMKK645 pKa = 10.39VGFYY649 pKa = 11.07ANDD652 pKa = 3.56SKK654 pKa = 11.01HH655 pKa = 4.33WQAVARR661 pKa = 11.84YY662 pKa = 7.13TSMLGVSEE670 pKa = 4.5TNDD673 pKa = 3.25VPLYY677 pKa = 10.23AANNEE682 pKa = 4.32MFSHH686 pKa = 6.7LTQSYY691 pKa = 9.58LQLNLTRR698 pKa = 11.84LFTMCAEE705 pKa = 4.34SVYY708 pKa = 10.42HH709 pKa = 7.28SNGTYY714 pKa = 10.09YY715 pKa = 11.09NPLTVVDD722 pKa = 4.39TACAITPVANSLHH735 pKa = 5.61TSIPAEE741 pKa = 3.8LLIIKK746 pKa = 7.91TKK748 pKa = 10.79AGAGTNVGDD757 pKa = 3.73QTVLGEE763 pKa = 4.68WPDD766 pKa = 3.86PWWDD770 pKa = 3.04WLLAGVRR777 pKa = 11.84AAIPFLGSSIPTAALVGAASGLHH800 pKa = 5.88SFLAEE805 pKa = 3.84KK806 pKa = 10.54LNPSRR811 pKa = 11.84AKK813 pKa = 9.81EE814 pKa = 3.85EE815 pKa = 3.86QMIARR820 pKa = 11.84AQATKK825 pKa = 10.56ALEE828 pKa = 4.16SSAEE832 pKa = 3.94AVHH835 pKa = 7.09DD836 pKa = 3.84VLSTGRR842 pKa = 11.84EE843 pKa = 3.72AVGRR847 pKa = 11.84ISKK850 pKa = 10.31AINDD854 pKa = 3.9FTSDD858 pKa = 3.54QLDD861 pKa = 3.68EE862 pKa = 4.26
Molecular weight: 95.87 kDa
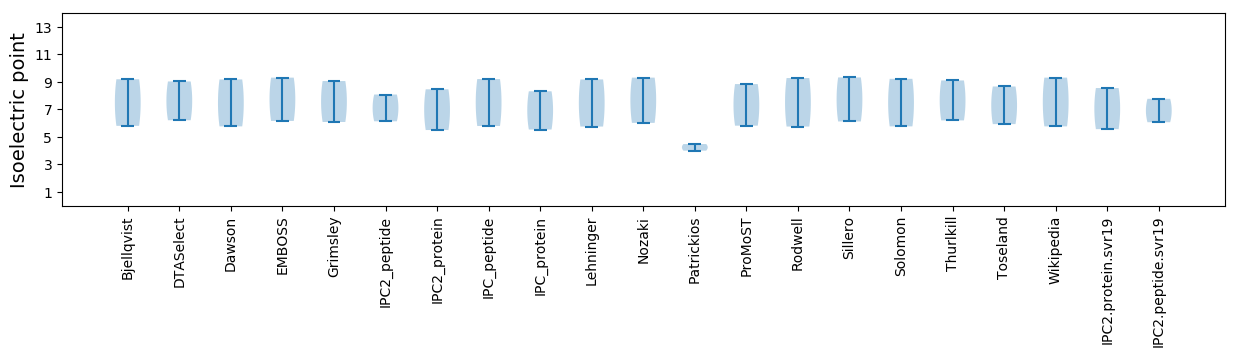
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KF57|A0A1L3KF57_9VIRU RdRp catalytic domain-containing protein OS=Hubei toti-like virus 12 OX=1923300 PE=4 SV=1
MM1 pKa = 7.22ARR3 pKa = 11.84SMVGLAVGWSACCNTLFGFKK23 pKa = 10.15HH24 pKa = 6.2PNGCTCRR31 pKa = 11.84CCEE34 pKa = 3.58WTALFFSGEE43 pKa = 4.12AKK45 pKa = 10.23PKK47 pKa = 8.02PCKK50 pKa = 10.02RR51 pKa = 11.84GANDD55 pKa = 3.33RR56 pKa = 11.84TGTSHH61 pKa = 6.9EE62 pKa = 4.43SAGVEE67 pKa = 4.12CGSCARR73 pKa = 11.84CVIDD77 pKa = 3.43RR78 pKa = 11.84TRR80 pKa = 11.84SSGTYY85 pKa = 8.91IKK87 pKa = 10.71SNQRR91 pKa = 11.84FYY93 pKa = 11.27EE94 pKa = 4.05RR95 pKa = 11.84STRR98 pKa = 11.84RR99 pKa = 11.84VTVSDD104 pKa = 3.79AQLPLYY110 pKa = 9.02WDD112 pKa = 3.47PGYY115 pKa = 9.91WVQAINLLQPASSSIEE131 pKa = 3.83FHH133 pKa = 7.77DD134 pKa = 4.78DD135 pKa = 3.23LSLHH139 pKa = 6.22RR140 pKa = 11.84SRR142 pKa = 11.84YY143 pKa = 8.93KK144 pKa = 9.57IHH146 pKa = 6.5SAPGVGVYY154 pKa = 9.83SRR156 pKa = 11.84PMSFYY161 pKa = 11.0LDD163 pKa = 3.2TLAQLEE169 pKa = 4.62VGCGIGSPYY178 pKa = 9.41WVLAVLLQRR187 pKa = 11.84ARR189 pKa = 11.84AAQDD193 pKa = 3.63PQLSCYY199 pKa = 8.94QWLKK203 pKa = 11.04GLTEE207 pKa = 5.11HH208 pKa = 7.11IEE210 pKa = 4.21TMDD213 pKa = 3.33GDD215 pKa = 4.06RR216 pKa = 11.84FVCSQILVGLEE227 pKa = 3.47RR228 pKa = 11.84VTPPKK233 pKa = 10.11QWSKK237 pKa = 11.91DD238 pKa = 3.62DD239 pKa = 5.07LIADD243 pKa = 4.39AKK245 pKa = 10.91NWVGPNKK252 pKa = 9.08TWASSRR258 pKa = 11.84GVRR261 pKa = 11.84EE262 pKa = 3.95WVEE265 pKa = 3.6GWVEE269 pKa = 3.94KK270 pKa = 10.81KK271 pKa = 9.29FAKK274 pKa = 8.62WQRR277 pKa = 11.84TMDD280 pKa = 3.59MIGRR284 pKa = 11.84EE285 pKa = 3.91NLSFEE290 pKa = 4.92EE291 pKa = 4.66FVQDD295 pKa = 3.38PTRR298 pKa = 11.84WATSGGAPRR307 pKa = 11.84VEE309 pKa = 4.21WEE311 pKa = 3.68GDD313 pKa = 3.19EE314 pKa = 5.08YY315 pKa = 10.76RR316 pKa = 11.84SKK318 pKa = 10.26WAWAMKK324 pKa = 10.07NISEE328 pKa = 4.25GKK330 pKa = 9.79YY331 pKa = 10.38LYY333 pKa = 10.66AQAKK337 pKa = 6.26TMPNIAHH344 pKa = 6.97AAIKK348 pKa = 9.96EE349 pKa = 4.32EE350 pKa = 4.1PSKK353 pKa = 10.12TRR355 pKa = 11.84LVITTPMSSYY365 pKa = 10.67LRR367 pKa = 11.84QSYY370 pKa = 10.05VMYY373 pKa = 10.77LFGIPEE379 pKa = 4.83LNSPIYY385 pKa = 10.62GNKK388 pKa = 9.15QLGEE392 pKa = 4.16LNNVKK397 pKa = 9.21WASYY401 pKa = 10.65CSIDD405 pKa = 4.58AKK407 pKa = 11.17AFDD410 pKa = 4.02HH411 pKa = 6.82QIPIWFIKK419 pKa = 10.53LVMEE423 pKa = 4.7HH424 pKa = 6.83LFKK427 pKa = 11.14SVGRR431 pKa = 11.84EE432 pKa = 3.81DD433 pKa = 2.9LWYY436 pKa = 10.37EE437 pKa = 3.75EE438 pKa = 4.12RR439 pKa = 11.84AHH441 pKa = 6.71INRR444 pKa = 11.84LKK446 pKa = 10.52IEE448 pKa = 4.19VFGEE452 pKa = 3.96IIPYY456 pKa = 9.82HH457 pKa = 6.22GGVLSGWRR465 pKa = 11.84LTSLLGTLASQALCEE480 pKa = 3.99WLILQGVNTNFVVQGDD496 pKa = 4.53DD497 pKa = 2.95ILLYY501 pKa = 10.19SAEE504 pKa = 4.85PIPPNVPDD512 pKa = 3.62MVQEE516 pKa = 4.61FGLKK520 pKa = 9.1IDD522 pKa = 4.16PVVSIPRR529 pKa = 11.84TCGYY533 pKa = 9.78FLRR536 pKa = 11.84RR537 pKa = 11.84AYY539 pKa = 10.25GGSFNLMSSGRR550 pKa = 11.84ALRR553 pKa = 11.84SLFYY557 pKa = 10.62ASPWVEE563 pKa = 3.21RR564 pKa = 11.84SQFQTPNSLARR575 pKa = 11.84SWLQFMSRR583 pKa = 11.84VPGKK587 pKa = 8.93SARR590 pKa = 11.84PWLLRR595 pKa = 11.84QAAADD600 pKa = 3.67MARR603 pKa = 11.84WARR606 pKa = 11.84WPGWTKK612 pKa = 10.65NRR614 pKa = 11.84WFEE617 pKa = 5.19LLTTDD622 pKa = 4.94AGLGGLGTVDD632 pKa = 2.68THH634 pKa = 7.76IYY636 pKa = 9.5RR637 pKa = 11.84QYY639 pKa = 11.02LPQIKK644 pKa = 10.0EE645 pKa = 4.07DD646 pKa = 3.44IDD648 pKa = 3.88RR649 pKa = 11.84SRR651 pKa = 11.84HH652 pKa = 5.87ARR654 pKa = 11.84TAWKK658 pKa = 9.96RR659 pKa = 11.84LYY661 pKa = 10.28EE662 pKa = 4.07YY663 pKa = 9.82FVPVSGLMAAKK674 pKa = 9.72DD675 pKa = 3.44QQVFYY680 pKa = 10.75RR681 pKa = 11.84MSYY684 pKa = 9.41RR685 pKa = 11.84PYY687 pKa = 10.57VRR689 pKa = 11.84PTPPTSAPKK698 pKa = 10.38LATDD702 pKa = 3.37WADD705 pKa = 3.42GVNKK709 pKa = 10.02TDD711 pKa = 2.9MCFNVMQGGFKK722 pKa = 10.72ALPEE726 pKa = 4.1DD727 pKa = 4.05VKK729 pKa = 11.02RR730 pKa = 11.84SAPHH734 pKa = 6.32WLRR737 pKa = 11.84TQPWFRR743 pKa = 11.84VLDD746 pKa = 3.45WLLRR750 pKa = 11.84PEE752 pKa = 4.72EE753 pKa = 4.1MTAPQHH759 pKa = 6.73LCIQPWVLNEE769 pKa = 4.04MLQSDD774 pKa = 4.0ISAADD779 pKa = 3.56RR780 pKa = 11.84VLHH783 pKa = 5.47NMKK786 pKa = 10.36KK787 pKa = 10.45GVISKK792 pKa = 9.96KK793 pKa = 9.33FSMYY797 pKa = 9.73QWILARR803 pKa = 11.84AGKK806 pKa = 10.54LMVALGSWW814 pKa = 3.25
MM1 pKa = 7.22ARR3 pKa = 11.84SMVGLAVGWSACCNTLFGFKK23 pKa = 10.15HH24 pKa = 6.2PNGCTCRR31 pKa = 11.84CCEE34 pKa = 3.58WTALFFSGEE43 pKa = 4.12AKK45 pKa = 10.23PKK47 pKa = 8.02PCKK50 pKa = 10.02RR51 pKa = 11.84GANDD55 pKa = 3.33RR56 pKa = 11.84TGTSHH61 pKa = 6.9EE62 pKa = 4.43SAGVEE67 pKa = 4.12CGSCARR73 pKa = 11.84CVIDD77 pKa = 3.43RR78 pKa = 11.84TRR80 pKa = 11.84SSGTYY85 pKa = 8.91IKK87 pKa = 10.71SNQRR91 pKa = 11.84FYY93 pKa = 11.27EE94 pKa = 4.05RR95 pKa = 11.84STRR98 pKa = 11.84RR99 pKa = 11.84VTVSDD104 pKa = 3.79AQLPLYY110 pKa = 9.02WDD112 pKa = 3.47PGYY115 pKa = 9.91WVQAINLLQPASSSIEE131 pKa = 3.83FHH133 pKa = 7.77DD134 pKa = 4.78DD135 pKa = 3.23LSLHH139 pKa = 6.22RR140 pKa = 11.84SRR142 pKa = 11.84YY143 pKa = 8.93KK144 pKa = 9.57IHH146 pKa = 6.5SAPGVGVYY154 pKa = 9.83SRR156 pKa = 11.84PMSFYY161 pKa = 11.0LDD163 pKa = 3.2TLAQLEE169 pKa = 4.62VGCGIGSPYY178 pKa = 9.41WVLAVLLQRR187 pKa = 11.84ARR189 pKa = 11.84AAQDD193 pKa = 3.63PQLSCYY199 pKa = 8.94QWLKK203 pKa = 11.04GLTEE207 pKa = 5.11HH208 pKa = 7.11IEE210 pKa = 4.21TMDD213 pKa = 3.33GDD215 pKa = 4.06RR216 pKa = 11.84FVCSQILVGLEE227 pKa = 3.47RR228 pKa = 11.84VTPPKK233 pKa = 10.11QWSKK237 pKa = 11.91DD238 pKa = 3.62DD239 pKa = 5.07LIADD243 pKa = 4.39AKK245 pKa = 10.91NWVGPNKK252 pKa = 9.08TWASSRR258 pKa = 11.84GVRR261 pKa = 11.84EE262 pKa = 3.95WVEE265 pKa = 3.6GWVEE269 pKa = 3.94KK270 pKa = 10.81KK271 pKa = 9.29FAKK274 pKa = 8.62WQRR277 pKa = 11.84TMDD280 pKa = 3.59MIGRR284 pKa = 11.84EE285 pKa = 3.91NLSFEE290 pKa = 4.92EE291 pKa = 4.66FVQDD295 pKa = 3.38PTRR298 pKa = 11.84WATSGGAPRR307 pKa = 11.84VEE309 pKa = 4.21WEE311 pKa = 3.68GDD313 pKa = 3.19EE314 pKa = 5.08YY315 pKa = 10.76RR316 pKa = 11.84SKK318 pKa = 10.26WAWAMKK324 pKa = 10.07NISEE328 pKa = 4.25GKK330 pKa = 9.79YY331 pKa = 10.38LYY333 pKa = 10.66AQAKK337 pKa = 6.26TMPNIAHH344 pKa = 6.97AAIKK348 pKa = 9.96EE349 pKa = 4.32EE350 pKa = 4.1PSKK353 pKa = 10.12TRR355 pKa = 11.84LVITTPMSSYY365 pKa = 10.67LRR367 pKa = 11.84QSYY370 pKa = 10.05VMYY373 pKa = 10.77LFGIPEE379 pKa = 4.83LNSPIYY385 pKa = 10.62GNKK388 pKa = 9.15QLGEE392 pKa = 4.16LNNVKK397 pKa = 9.21WASYY401 pKa = 10.65CSIDD405 pKa = 4.58AKK407 pKa = 11.17AFDD410 pKa = 4.02HH411 pKa = 6.82QIPIWFIKK419 pKa = 10.53LVMEE423 pKa = 4.7HH424 pKa = 6.83LFKK427 pKa = 11.14SVGRR431 pKa = 11.84EE432 pKa = 3.81DD433 pKa = 2.9LWYY436 pKa = 10.37EE437 pKa = 3.75EE438 pKa = 4.12RR439 pKa = 11.84AHH441 pKa = 6.71INRR444 pKa = 11.84LKK446 pKa = 10.52IEE448 pKa = 4.19VFGEE452 pKa = 3.96IIPYY456 pKa = 9.82HH457 pKa = 6.22GGVLSGWRR465 pKa = 11.84LTSLLGTLASQALCEE480 pKa = 3.99WLILQGVNTNFVVQGDD496 pKa = 4.53DD497 pKa = 2.95ILLYY501 pKa = 10.19SAEE504 pKa = 4.85PIPPNVPDD512 pKa = 3.62MVQEE516 pKa = 4.61FGLKK520 pKa = 9.1IDD522 pKa = 4.16PVVSIPRR529 pKa = 11.84TCGYY533 pKa = 9.78FLRR536 pKa = 11.84RR537 pKa = 11.84AYY539 pKa = 10.25GGSFNLMSSGRR550 pKa = 11.84ALRR553 pKa = 11.84SLFYY557 pKa = 10.62ASPWVEE563 pKa = 3.21RR564 pKa = 11.84SQFQTPNSLARR575 pKa = 11.84SWLQFMSRR583 pKa = 11.84VPGKK587 pKa = 8.93SARR590 pKa = 11.84PWLLRR595 pKa = 11.84QAAADD600 pKa = 3.67MARR603 pKa = 11.84WARR606 pKa = 11.84WPGWTKK612 pKa = 10.65NRR614 pKa = 11.84WFEE617 pKa = 5.19LLTTDD622 pKa = 4.94AGLGGLGTVDD632 pKa = 2.68THH634 pKa = 7.76IYY636 pKa = 9.5RR637 pKa = 11.84QYY639 pKa = 11.02LPQIKK644 pKa = 10.0EE645 pKa = 4.07DD646 pKa = 3.44IDD648 pKa = 3.88RR649 pKa = 11.84SRR651 pKa = 11.84HH652 pKa = 5.87ARR654 pKa = 11.84TAWKK658 pKa = 9.96RR659 pKa = 11.84LYY661 pKa = 10.28EE662 pKa = 4.07YY663 pKa = 9.82FVPVSGLMAAKK674 pKa = 9.72DD675 pKa = 3.44QQVFYY680 pKa = 10.75RR681 pKa = 11.84MSYY684 pKa = 9.41RR685 pKa = 11.84PYY687 pKa = 10.57VRR689 pKa = 11.84PTPPTSAPKK698 pKa = 10.38LATDD702 pKa = 3.37WADD705 pKa = 3.42GVNKK709 pKa = 10.02TDD711 pKa = 2.9MCFNVMQGGFKK722 pKa = 10.72ALPEE726 pKa = 4.1DD727 pKa = 4.05VKK729 pKa = 11.02RR730 pKa = 11.84SAPHH734 pKa = 6.32WLRR737 pKa = 11.84TQPWFRR743 pKa = 11.84VLDD746 pKa = 3.45WLLRR750 pKa = 11.84PEE752 pKa = 4.72EE753 pKa = 4.1MTAPQHH759 pKa = 6.73LCIQPWVLNEE769 pKa = 4.04MLQSDD774 pKa = 4.0ISAADD779 pKa = 3.56RR780 pKa = 11.84VLHH783 pKa = 5.47NMKK786 pKa = 10.36KK787 pKa = 10.45GVISKK792 pKa = 9.96KK793 pKa = 9.33FSMYY797 pKa = 9.73QWILARR803 pKa = 11.84AGKK806 pKa = 10.54LMVALGSWW814 pKa = 3.25
Molecular weight: 93.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1676 |
814 |
862 |
838.0 |
94.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.413 ± 0.472 | 1.85 ± 0.254 |
4.714 ± 0.204 | 4.952 ± 0.059 |
2.924 ± 0.534 | 6.086 ± 0.557 |
2.387 ± 0.295 | 4.654 ± 0.162 |
4.057 ± 0.687 | 9.01 ± 0.115 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.341 ± 0.275 | 3.938 ± 0.608 |
5.668 ± 0.012 | 4.296 ± 0.003 |
5.907 ± 0.854 | 8.234 ± 0.433 |
5.131 ± 0.325 | 7.279 ± 0.797 |
3.938 ± 0.34 | 3.222 ± 0.411 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
