
Microviridae Fen418_41
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
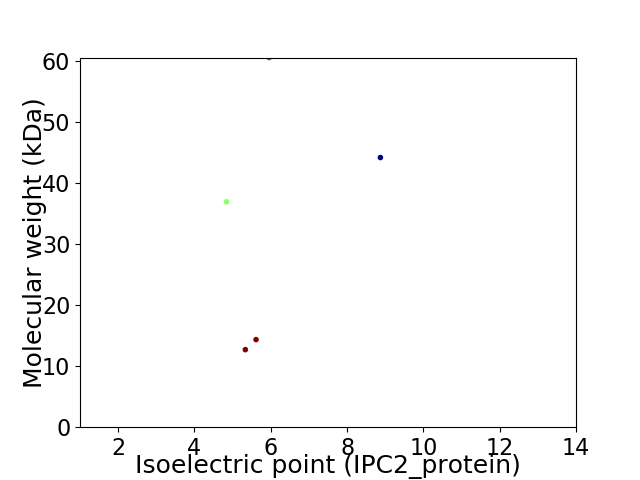
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G2UME7|A0A0G2UME7_9VIRU Uncharacterized protein OS=Microviridae Fen418_41 OX=1655653 PE=4 SV=1
MM1 pKa = 7.21WRR3 pKa = 11.84HH4 pKa = 5.51GVAIMIIVYY13 pKa = 10.09DD14 pKa = 3.76HH15 pKa = 7.31LIFLSKK21 pKa = 9.88RR22 pKa = 11.84TMVFLFIFLQTTAAGGGISVLGALAGAIPMVGGIISGLINNSNQDD67 pKa = 3.04QVNASNQAFQLAQQQQIEE85 pKa = 4.76AYY87 pKa = 8.45NTAQWNAQNAYY98 pKa = 9.79NSPEE102 pKa = 3.91EE103 pKa = 4.0VMQRR107 pKa = 11.84YY108 pKa = 8.82EE109 pKa = 4.66DD110 pKa = 4.1AGLNPNLIYY119 pKa = 10.76GSGSGTGNLAEE130 pKa = 4.63MPQPAPRR137 pKa = 11.84VEE139 pKa = 5.21FVAQAHH145 pKa = 6.51PNFDD149 pKa = 3.97LGTAAAQSVQAYY161 pKa = 9.02NNTVSTVNQGNLVKK175 pKa = 10.51QQSINVAQNTALQAAQEE192 pKa = 4.17ININADD198 pKa = 2.98TGLKK202 pKa = 10.38DD203 pKa = 3.62NQEE206 pKa = 4.03AAQLISNLYY215 pKa = 10.51SGDD218 pKa = 3.59SMKK221 pKa = 11.3ANIALMKK228 pKa = 10.56AQTQLVLTNQQTAQLMQQPNLVTALQNITNMQEE261 pKa = 3.94NNQLTQAQIANINANIAQTQIKK283 pKa = 8.59TKK285 pKa = 10.54LDD287 pKa = 3.41QLNLYY292 pKa = 8.48LQNHH296 pKa = 6.46GLNLDD301 pKa = 3.64SSLMSRR307 pKa = 11.84VGSIVLQNLSKK318 pKa = 10.24STVASKK324 pKa = 10.46MNSADD329 pKa = 3.7NSPDD333 pKa = 3.45MNDD336 pKa = 2.81FDD338 pKa = 5.08YY339 pKa = 11.54NYY341 pKa = 11.38
MM1 pKa = 7.21WRR3 pKa = 11.84HH4 pKa = 5.51GVAIMIIVYY13 pKa = 10.09DD14 pKa = 3.76HH15 pKa = 7.31LIFLSKK21 pKa = 9.88RR22 pKa = 11.84TMVFLFIFLQTTAAGGGISVLGALAGAIPMVGGIISGLINNSNQDD67 pKa = 3.04QVNASNQAFQLAQQQQIEE85 pKa = 4.76AYY87 pKa = 8.45NTAQWNAQNAYY98 pKa = 9.79NSPEE102 pKa = 3.91EE103 pKa = 4.0VMQRR107 pKa = 11.84YY108 pKa = 8.82EE109 pKa = 4.66DD110 pKa = 4.1AGLNPNLIYY119 pKa = 10.76GSGSGTGNLAEE130 pKa = 4.63MPQPAPRR137 pKa = 11.84VEE139 pKa = 5.21FVAQAHH145 pKa = 6.51PNFDD149 pKa = 3.97LGTAAAQSVQAYY161 pKa = 9.02NNTVSTVNQGNLVKK175 pKa = 10.51QQSINVAQNTALQAAQEE192 pKa = 4.17ININADD198 pKa = 2.98TGLKK202 pKa = 10.38DD203 pKa = 3.62NQEE206 pKa = 4.03AAQLISNLYY215 pKa = 10.51SGDD218 pKa = 3.59SMKK221 pKa = 11.3ANIALMKK228 pKa = 10.56AQTQLVLTNQQTAQLMQQPNLVTALQNITNMQEE261 pKa = 3.94NNQLTQAQIANINANIAQTQIKK283 pKa = 8.59TKK285 pKa = 10.54LDD287 pKa = 3.41QLNLYY292 pKa = 8.48LQNHH296 pKa = 6.46GLNLDD301 pKa = 3.64SSLMSRR307 pKa = 11.84VGSIVLQNLSKK318 pKa = 10.24STVASKK324 pKa = 10.46MNSADD329 pKa = 3.7NSPDD333 pKa = 3.45MNDD336 pKa = 2.81FDD338 pKa = 5.08YY339 pKa = 11.54NYY341 pKa = 11.38
Molecular weight: 36.98 kDa
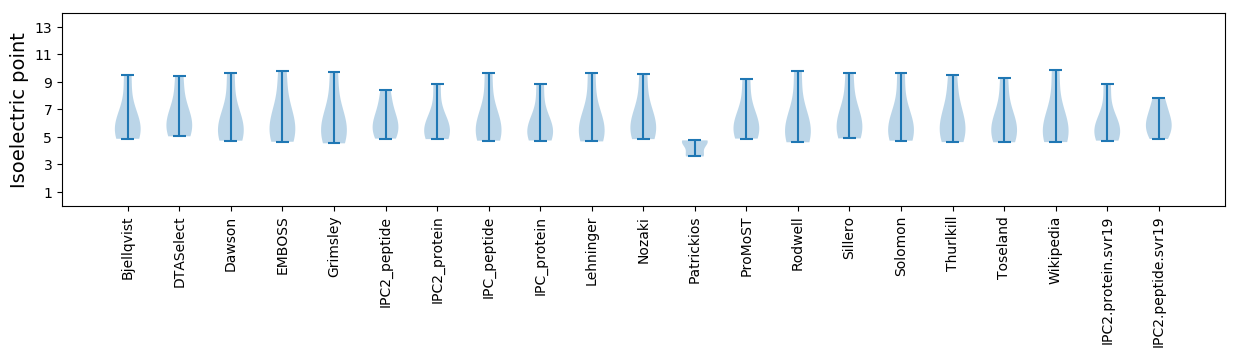
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G2UK30|A0A0G2UK30_9VIRU DNA pilot protein VP2 OS=Microviridae Fen418_41 OX=1655653 PE=4 SV=1
MM1 pKa = 7.47IVFLGLISCGSISFLTLFLLITPVIVLTLLIRR33 pKa = 11.84SFLSLGVIMTVGLTSLFALSGRR55 pKa = 11.84KK56 pKa = 9.02KK57 pKa = 10.3KK58 pKa = 11.01YY59 pKa = 9.01EE60 pKa = 3.74LSFSIMAKK68 pKa = 8.39KK69 pKa = 8.7TLINMQQITNIYY81 pKa = 7.75MPCRR85 pKa = 11.84FPFQTKK91 pKa = 9.97DD92 pKa = 3.28GAIVPCGKK100 pKa = 9.97CDD102 pKa = 3.32SCLTRR107 pKa = 11.84MANGWAFRR115 pKa = 11.84VMQEE119 pKa = 3.89EE120 pKa = 4.46KK121 pKa = 10.38KK122 pKa = 10.35AKK124 pKa = 9.08CSMWITLTYY133 pKa = 9.85DD134 pKa = 3.25TDD136 pKa = 4.3HH137 pKa = 7.2VPLTNNGYY145 pKa = 7.1MTLNRR150 pKa = 11.84VHH152 pKa = 6.05TQLFFKK158 pKa = 10.67RR159 pKa = 11.84LRR161 pKa = 11.84RR162 pKa = 11.84FQNIHH167 pKa = 6.68NINQEE172 pKa = 4.2RR173 pKa = 11.84IYY175 pKa = 10.63DD176 pKa = 3.52TGDD179 pKa = 2.84RR180 pKa = 11.84FGRR183 pKa = 11.84LQYY186 pKa = 9.27PIKK189 pKa = 10.76YY190 pKa = 9.16FGCGEE195 pKa = 4.19YY196 pKa = 8.89GTKK199 pKa = 9.82YY200 pKa = 10.09RR201 pKa = 11.84RR202 pKa = 11.84PHH204 pKa = 4.56YY205 pKa = 9.79HH206 pKa = 6.56ICLLNCDD213 pKa = 5.03EE214 pKa = 4.22RR215 pKa = 11.84ALWYY219 pKa = 10.03SWQDD223 pKa = 3.22GEE225 pKa = 4.53TGLPFGDD232 pKa = 4.25VYY234 pKa = 11.04IDD236 pKa = 4.6DD237 pKa = 4.9RR238 pKa = 11.84PLSAAAIAYY247 pKa = 7.11TCFYY251 pKa = 9.01MNKK254 pKa = 8.23PKK256 pKa = 10.36RR257 pKa = 11.84VPEE260 pKa = 4.09HH261 pKa = 6.78KK262 pKa = 10.52NDD264 pKa = 3.23DD265 pKa = 3.68RR266 pKa = 11.84EE267 pKa = 4.21RR268 pKa = 11.84EE269 pKa = 3.87FRR271 pKa = 11.84MMSTKK276 pKa = 9.91MGLGYY281 pKa = 8.37LTPDD285 pKa = 2.91TLAYY289 pKa = 10.16HH290 pKa = 6.69KK291 pKa = 10.94ADD293 pKa = 3.61LSRR296 pKa = 11.84SYY298 pKa = 9.96FTTEE302 pKa = 3.8GGGKK306 pKa = 9.72AALPRR311 pKa = 11.84YY312 pKa = 9.12LADD315 pKa = 5.24RR316 pKa = 11.84IFTPEE321 pKa = 4.1EE322 pKa = 3.92KK323 pKa = 10.43ASQQPLINEE332 pKa = 4.14RR333 pKa = 11.84NSLQFNLEE341 pKa = 3.55FDD343 pKa = 3.35EE344 pKa = 4.64HH345 pKa = 6.19KK346 pKa = 10.69RR347 pKa = 11.84RR348 pKa = 11.84TGGNRR353 pKa = 11.84QTFQQEE359 pKa = 4.02TNDD362 pKa = 3.33RR363 pKa = 11.84RR364 pKa = 11.84RR365 pKa = 11.84EE366 pKa = 4.2SIRR369 pKa = 11.84SFQRR373 pKa = 11.84KK374 pKa = 8.43ARR376 pKa = 11.84PDD378 pKa = 3.21GMM380 pKa = 4.86
MM1 pKa = 7.47IVFLGLISCGSISFLTLFLLITPVIVLTLLIRR33 pKa = 11.84SFLSLGVIMTVGLTSLFALSGRR55 pKa = 11.84KK56 pKa = 9.02KK57 pKa = 10.3KK58 pKa = 11.01YY59 pKa = 9.01EE60 pKa = 3.74LSFSIMAKK68 pKa = 8.39KK69 pKa = 8.7TLINMQQITNIYY81 pKa = 7.75MPCRR85 pKa = 11.84FPFQTKK91 pKa = 9.97DD92 pKa = 3.28GAIVPCGKK100 pKa = 9.97CDD102 pKa = 3.32SCLTRR107 pKa = 11.84MANGWAFRR115 pKa = 11.84VMQEE119 pKa = 3.89EE120 pKa = 4.46KK121 pKa = 10.38KK122 pKa = 10.35AKK124 pKa = 9.08CSMWITLTYY133 pKa = 9.85DD134 pKa = 3.25TDD136 pKa = 4.3HH137 pKa = 7.2VPLTNNGYY145 pKa = 7.1MTLNRR150 pKa = 11.84VHH152 pKa = 6.05TQLFFKK158 pKa = 10.67RR159 pKa = 11.84LRR161 pKa = 11.84RR162 pKa = 11.84FQNIHH167 pKa = 6.68NINQEE172 pKa = 4.2RR173 pKa = 11.84IYY175 pKa = 10.63DD176 pKa = 3.52TGDD179 pKa = 2.84RR180 pKa = 11.84FGRR183 pKa = 11.84LQYY186 pKa = 9.27PIKK189 pKa = 10.76YY190 pKa = 9.16FGCGEE195 pKa = 4.19YY196 pKa = 8.89GTKK199 pKa = 9.82YY200 pKa = 10.09RR201 pKa = 11.84RR202 pKa = 11.84PHH204 pKa = 4.56YY205 pKa = 9.79HH206 pKa = 6.56ICLLNCDD213 pKa = 5.03EE214 pKa = 4.22RR215 pKa = 11.84ALWYY219 pKa = 10.03SWQDD223 pKa = 3.22GEE225 pKa = 4.53TGLPFGDD232 pKa = 4.25VYY234 pKa = 11.04IDD236 pKa = 4.6DD237 pKa = 4.9RR238 pKa = 11.84PLSAAAIAYY247 pKa = 7.11TCFYY251 pKa = 9.01MNKK254 pKa = 8.23PKK256 pKa = 10.36RR257 pKa = 11.84VPEE260 pKa = 4.09HH261 pKa = 6.78KK262 pKa = 10.52NDD264 pKa = 3.23DD265 pKa = 3.68RR266 pKa = 11.84EE267 pKa = 4.21RR268 pKa = 11.84EE269 pKa = 3.87FRR271 pKa = 11.84MMSTKK276 pKa = 9.91MGLGYY281 pKa = 8.37LTPDD285 pKa = 2.91TLAYY289 pKa = 10.16HH290 pKa = 6.69KK291 pKa = 10.94ADD293 pKa = 3.61LSRR296 pKa = 11.84SYY298 pKa = 9.96FTTEE302 pKa = 3.8GGGKK306 pKa = 9.72AALPRR311 pKa = 11.84YY312 pKa = 9.12LADD315 pKa = 5.24RR316 pKa = 11.84IFTPEE321 pKa = 4.1EE322 pKa = 3.92KK323 pKa = 10.43ASQQPLINEE332 pKa = 4.14RR333 pKa = 11.84NSLQFNLEE341 pKa = 3.55FDD343 pKa = 3.35EE344 pKa = 4.64HH345 pKa = 6.19KK346 pKa = 10.69RR347 pKa = 11.84RR348 pKa = 11.84TGGNRR353 pKa = 11.84QTFQQEE359 pKa = 4.02TNDD362 pKa = 3.33RR363 pKa = 11.84RR364 pKa = 11.84RR365 pKa = 11.84EE366 pKa = 4.2SIRR369 pKa = 11.84SFQRR373 pKa = 11.84KK374 pKa = 8.43ARR376 pKa = 11.84PDD378 pKa = 3.21GMM380 pKa = 4.86
Molecular weight: 44.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1508 |
115 |
551 |
301.6 |
33.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.891 ± 1.228 | 1.26 ± 0.481 |
4.841 ± 0.958 | 4.31 ± 0.673 |
4.31 ± 0.698 | 6.83 ± 1.115 |
1.923 ± 0.238 | 6.366 ± 0.168 |
3.581 ± 0.908 | 8.554 ± 0.74 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.249 ± 0.334 | 6.83 ± 1.483 |
4.973 ± 0.954 | 6.3 ± 1.528 |
5.637 ± 1.496 | 6.101 ± 0.356 |
7.029 ± 0.812 | 5.106 ± 0.762 |
1.393 ± 0.451 | 3.515 ± 0.475 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
