
Parvibaculum lavamentivorans (strain DS-1 / DSM 13023 / NCIMB 13966)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Parvibaculaceae; Parvibaculum; Parvibaculum lavamentivorans
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
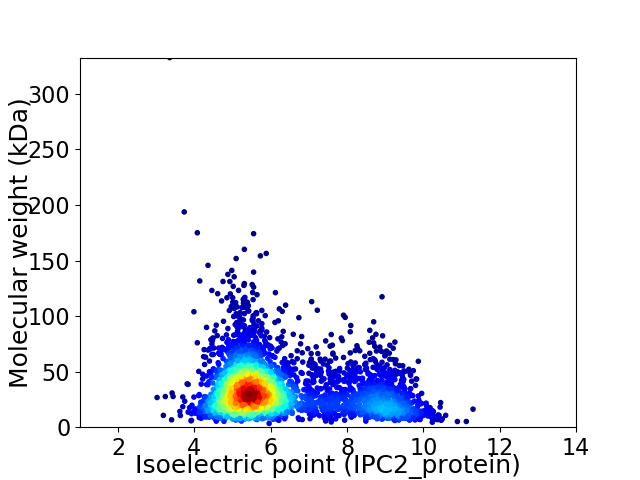
Virtual 2D-PAGE plot for 3580 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|A7HTA7|Y1521_PARL1 UPF0178 protein Plav_1521 OS=Parvibaculum lavamentivorans (strain DS-1 / DSM 13023 / NCIMB 13966) OX=402881 GN=Plav_1521 PE=3 SV=1
MM1 pKa = 6.53TTRR4 pKa = 11.84HH5 pKa = 6.0SKK7 pKa = 10.87NITGTTGAIDD17 pKa = 3.58SSLLGSAYY25 pKa = 10.15SYY27 pKa = 11.25YY28 pKa = 10.94GGVVQTAHH36 pKa = 6.75SSGKK40 pKa = 7.27TASGVKK46 pKa = 9.76RR47 pKa = 11.84VFAAAMLSGASVLAVGIAGTGNAAAGTCVGPINEE81 pKa = 4.91TYY83 pKa = 9.52TCAGTFDD90 pKa = 3.9DD91 pKa = 4.81TIEE94 pKa = 4.24YY95 pKa = 10.57DD96 pKa = 3.56SGVDD100 pKa = 4.48DD101 pKa = 3.77VTLVLGPGSFIDD113 pKa = 3.62TTEE116 pKa = 3.86NTSEE120 pKa = 4.43HH121 pKa = 6.49DD122 pKa = 3.46HH123 pKa = 7.11DD124 pKa = 3.99NAGIVVVGEE133 pKa = 3.82SDD135 pKa = 3.34MAIVNNGSIFTGDD148 pKa = 2.73NTTWVEE154 pKa = 3.59TDD156 pKa = 3.15EE157 pKa = 4.59EE158 pKa = 4.44YY159 pKa = 10.99GYY161 pKa = 10.88YY162 pKa = 10.42AYY164 pKa = 10.88LGDD167 pKa = 3.89GHH169 pKa = 8.13HH170 pKa = 7.29GIAAYY175 pKa = 10.68SNWDD179 pKa = 3.45DD180 pKa = 3.74AAVEE184 pKa = 4.06NTAFGTIVTTSSEE197 pKa = 4.21SHH199 pKa = 5.85GMVAEE204 pKa = 4.1ALEE207 pKa = 4.94GSASADD213 pKa = 3.29NAGLIGTSGEE223 pKa = 4.13DD224 pKa = 3.1SHH226 pKa = 7.73GISATAKK233 pKa = 8.98YY234 pKa = 10.52SVDD237 pKa = 3.37VVNSGTITTDD247 pKa = 2.61GTYY250 pKa = 10.75SHH252 pKa = 7.26GIWAALDD259 pKa = 3.4SEE261 pKa = 4.52YY262 pKa = 10.93GYY264 pKa = 11.47GSFGGIIEE272 pKa = 4.34ISNSGSIEE280 pKa = 4.02TSGEE284 pKa = 4.13SASGISANAEE294 pKa = 3.89EE295 pKa = 5.1GDD297 pKa = 3.93VEE299 pKa = 4.31IANSGSIVTEE309 pKa = 3.78GDD311 pKa = 3.05EE312 pKa = 4.93ALGIWAHH319 pKa = 6.44GDD321 pKa = 3.43DD322 pKa = 3.74VDD324 pKa = 5.16VVNTADD330 pKa = 3.89GSIVTYY336 pKa = 10.26GADD339 pKa = 2.93AHH341 pKa = 6.06GVYY344 pKa = 10.3IFGDD348 pKa = 3.87TVSLSNAGTIEE359 pKa = 4.23TYY361 pKa = 10.98GEE363 pKa = 3.92DD364 pKa = 3.02AHH366 pKa = 8.29AVVAYY371 pKa = 10.3SGGIDD376 pKa = 3.14TTTIVNTGLIQATGEE391 pKa = 3.82EE392 pKa = 4.17ADD394 pKa = 4.7AIRR397 pKa = 11.84ASGPTVRR404 pKa = 11.84ITNDD408 pKa = 3.01VIEE411 pKa = 4.51EE412 pKa = 4.12DD413 pKa = 4.35EE414 pKa = 4.32EE415 pKa = 4.57VVDD418 pKa = 3.78YY419 pKa = 11.18GVIRR423 pKa = 11.84SEE425 pKa = 4.34DD426 pKa = 3.2GAAIHH431 pKa = 5.6VTEE434 pKa = 5.22ADD436 pKa = 4.07DD437 pKa = 4.5ARR439 pKa = 11.84LYY441 pKa = 11.41NNGAIYY447 pKa = 10.58GNVSIEE453 pKa = 3.94TDD455 pKa = 3.2EE456 pKa = 4.04YY457 pKa = 11.09GYY459 pKa = 11.42AVNTGSISSDD469 pKa = 3.56RR470 pKa = 11.84KK471 pKa = 8.89WKK473 pKa = 10.26AALAIDD479 pKa = 4.14AEE481 pKa = 4.41EE482 pKa = 5.05GDD484 pKa = 4.02VVVVNDD490 pKa = 4.19GDD492 pKa = 3.89IEE494 pKa = 4.4ASGRR498 pKa = 11.84KK499 pKa = 9.17AKK501 pKa = 10.35GVEE504 pKa = 3.84LEE506 pKa = 4.76GYY508 pKa = 10.32DD509 pKa = 3.51VTLTNTGSIEE519 pKa = 4.0TWGSDD524 pKa = 2.74GHH526 pKa = 7.82AVDD529 pKa = 5.38LEE531 pKa = 4.05GDD533 pKa = 3.67EE534 pKa = 4.91EE535 pKa = 4.52YY536 pKa = 11.63GEE538 pKa = 4.31VTLNNSGLIQASGEE552 pKa = 3.9EE553 pKa = 4.04ADD555 pKa = 3.81AVRR558 pKa = 11.84ASGEE562 pKa = 4.23TVRR565 pKa = 11.84ITNQAVLVPGEE576 pKa = 3.91TDD578 pKa = 3.64AYY580 pKa = 9.93DD581 pKa = 3.35TTIYY585 pKa = 10.69GIIRR589 pKa = 11.84SEE591 pKa = 4.17DD592 pKa = 3.16GAAIRR597 pKa = 11.84VDD599 pKa = 3.64EE600 pKa = 4.44SDD602 pKa = 3.18VVYY605 pKa = 10.79VVNDD609 pKa = 3.17GFIYY613 pKa = 10.72SNISIEE619 pKa = 4.33ADD621 pKa = 3.19DD622 pKa = 3.81YY623 pKa = 11.83AYY625 pKa = 11.1VSNAGSVSSDD635 pKa = 3.44RR636 pKa = 11.84RR637 pKa = 11.84WKK639 pKa = 10.41AAVSADD645 pKa = 3.67VEE647 pKa = 4.39EE648 pKa = 4.96GNAVIVNDD656 pKa = 4.22GEE658 pKa = 4.49IVTTASRR665 pKa = 11.84AAGIEE670 pKa = 3.9VDD672 pKa = 4.03VEE674 pKa = 4.07LGDD677 pKa = 4.07AYY679 pKa = 11.11VVNSGTITTGSLEE692 pKa = 4.04EE693 pKa = 4.61GEE695 pKa = 4.81PEE697 pKa = 4.56GGWRR701 pKa = 11.84SHH703 pKa = 6.74GIDD706 pKa = 3.44VFAEE710 pKa = 4.03DD711 pKa = 3.99TVLVLNTGDD720 pKa = 3.33VTTYY724 pKa = 8.85GHH726 pKa = 6.64KK727 pKa = 10.74AKK729 pKa = 10.61GIRR732 pKa = 11.84IEE734 pKa = 4.3TQEE737 pKa = 3.74GTAAGVNSGTIDD749 pKa = 2.92TWGEE753 pKa = 3.86DD754 pKa = 3.42STGLIVTATSRR765 pKa = 11.84EE766 pKa = 4.23HH767 pKa = 6.32YY768 pKa = 10.29DD769 pKa = 3.27GYY771 pKa = 11.28SYY773 pKa = 11.57FDD775 pKa = 3.12ISGIALAGNIGSVEE789 pKa = 4.07TRR791 pKa = 11.84GDD793 pKa = 3.4DD794 pKa = 3.36AMGVVAVAEE803 pKa = 4.64GNFAAALNVLGGSISTSGDD822 pKa = 3.37DD823 pKa = 3.37AHH825 pKa = 7.63GLVAVSGFEE834 pKa = 4.15SVEE837 pKa = 3.99DD838 pKa = 3.65ALEE841 pKa = 4.2GSEE844 pKa = 3.97YY845 pKa = 10.98SEE847 pKa = 4.64GGEE850 pKa = 3.92GSYY853 pKa = 10.9AIAVNGLPPQLLAGFSGIGTEE874 pKa = 4.1MGSGVLGEE882 pKa = 4.29IADD885 pKa = 4.14VLAEE889 pKa = 4.15ALPEE893 pKa = 4.27GEE895 pKa = 4.55YY896 pKa = 11.11DD897 pKa = 3.58PADD900 pKa = 3.62FRR902 pKa = 11.84STIVTTGDD910 pKa = 3.18GAIGVLAVSSNGGAVAGNLYY930 pKa = 10.48GDD932 pKa = 3.81VVTGVLHH939 pKa = 6.16EE940 pKa = 4.66EE941 pKa = 5.26GYY943 pKa = 10.78SVGGDD948 pKa = 3.28HH949 pKa = 7.46AYY951 pKa = 10.22GVAAMSVDD959 pKa = 3.42EE960 pKa = 4.88GGAIAFNAYY969 pKa = 9.23NASIVTNGAYY979 pKa = 10.18AHH981 pKa = 6.65GLLAISEE988 pKa = 5.08DD989 pKa = 3.8GDD991 pKa = 3.56AMAMNKK997 pKa = 9.37YY998 pKa = 10.26ASSIVTNGLGAVGIRR1013 pKa = 11.84VWTDD1017 pKa = 3.27GEE1019 pKa = 4.07SDD1021 pKa = 3.85EE1022 pKa = 4.87YY1023 pKa = 11.61SSADD1027 pKa = 3.28ALVWNVGSSVTTYY1040 pKa = 10.72GQGAQGIVAFSEE1052 pKa = 4.17EE1053 pKa = 4.25GDD1055 pKa = 3.72VTVANAPMTIGEE1067 pKa = 4.55GEE1069 pKa = 4.33DD1070 pKa = 3.53EE1071 pKa = 4.97EE1072 pKa = 4.65ILAGLIEE1079 pKa = 4.29THH1081 pKa = 6.64GDD1083 pKa = 3.28EE1084 pKa = 5.88ADD1086 pKa = 4.67GIWASGEE1093 pKa = 4.06SDD1095 pKa = 3.04VDD1097 pKa = 4.07VYY1099 pKa = 11.82NSGSIITHH1107 pKa = 5.56GEE1109 pKa = 3.83GANGISASGDD1119 pKa = 3.3EE1120 pKa = 4.07VDD1122 pKa = 4.55IDD1124 pKa = 3.81NSGTIQTSGDD1134 pKa = 3.37EE1135 pKa = 4.16AFGVDD1140 pKa = 3.36MRR1142 pKa = 11.84GWSVSLDD1149 pKa = 3.4NSGTISTSGKK1159 pKa = 8.38YY1160 pKa = 9.2AHH1162 pKa = 6.92AVVGDD1167 pKa = 4.17SYY1169 pKa = 12.15SDD1171 pKa = 3.46LTTTVYY1177 pKa = 11.08NSGTIEE1183 pKa = 4.04ATGKK1187 pKa = 9.1YY1188 pKa = 9.47ANAVRR1193 pKa = 11.84VSGPTVHH1200 pKa = 5.76VTNTEE1205 pKa = 4.02DD1206 pKa = 3.71GVISSGDD1213 pKa = 3.28VAIYY1217 pKa = 10.62AFEE1220 pKa = 4.08TKK1222 pKa = 9.91YY1223 pKa = 11.42ASIANHH1229 pKa = 6.26GEE1231 pKa = 4.17VEE1233 pKa = 4.29GSIHH1237 pKa = 6.13VSAYY1241 pKa = 10.32DD1242 pKa = 3.73YY1243 pKa = 11.55EE1244 pKa = 5.65DD1245 pKa = 3.57FASTYY1250 pKa = 9.94IVNTGSVSGDD1260 pKa = 2.56IDD1262 pKa = 3.76TSFGEE1267 pKa = 4.08SDD1269 pKa = 4.95DD1270 pKa = 4.46IIIVDD1275 pKa = 4.08GGTVGGAIFTGDD1287 pKa = 3.59GEE1289 pKa = 4.79DD1290 pKa = 3.49EE1291 pKa = 4.3VTISGTGVSIAKK1303 pKa = 9.79GIHH1306 pKa = 6.21GDD1308 pKa = 3.88GEE1310 pKa = 4.65GDD1312 pKa = 3.4LHH1314 pKa = 8.15VLFEE1318 pKa = 4.31QDD1320 pKa = 3.25DD1321 pKa = 4.39TVTFSDD1327 pKa = 4.85GIEE1330 pKa = 4.63GYY1332 pKa = 10.5AISNAHH1338 pKa = 5.75FVGFSSGTTVFDD1350 pKa = 3.58GVNIHH1355 pKa = 5.67TEE1357 pKa = 3.72EE1358 pKa = 5.47GEE1360 pKa = 4.07ILVGEE1365 pKa = 4.59DD1366 pKa = 2.86ATLATTIEE1374 pKa = 4.14NAFAFADD1381 pKa = 3.87EE1382 pKa = 4.34TSVLGRR1388 pKa = 11.84LKK1390 pKa = 10.7VVAGSSFGFSGDD1402 pKa = 3.55VVFDD1406 pKa = 3.57EE1407 pKa = 5.38GGTFEE1412 pKa = 4.77TGLTGDD1418 pKa = 3.63GAGVVTGDD1426 pKa = 3.51TVRR1429 pKa = 11.84FASGSTIHH1437 pKa = 6.48VDD1439 pKa = 3.02AGAGFTEE1446 pKa = 4.79TVGSDD1451 pKa = 3.24FLIASSEE1458 pKa = 4.33SEE1460 pKa = 4.44SGVTDD1465 pKa = 4.24DD1466 pKa = 5.08GASVTDD1472 pKa = 3.58NLILFKK1478 pKa = 10.45FLKK1481 pKa = 10.28VMNGDD1486 pKa = 3.76VVSEE1490 pKa = 4.41GAADD1494 pKa = 3.78EE1495 pKa = 4.3LFLRR1499 pKa = 11.84IEE1501 pKa = 4.19VEE1503 pKa = 3.93EE1504 pKa = 4.18TAFDD1508 pKa = 4.78LEE1510 pKa = 4.63TEE1512 pKa = 4.3AKK1514 pKa = 9.14GTGNLLSVASALDD1527 pKa = 3.82VYY1529 pKa = 11.07IEE1531 pKa = 4.21TQPLDD1536 pKa = 3.78NPLVQYY1542 pKa = 10.64LLQFEE1547 pKa = 4.66TEE1549 pKa = 4.26EE1550 pKa = 4.07EE1551 pKa = 3.88QLAALLKK1558 pKa = 10.11VIKK1561 pKa = 9.66DD1562 pKa = 3.85TVPDD1566 pKa = 3.87EE1567 pKa = 4.83SNASAGATIASTDD1580 pKa = 4.46LIYY1583 pKa = 11.38DD1584 pKa = 4.05MIMDD1588 pKa = 4.84RR1589 pKa = 11.84LSGGGFVVADD1599 pKa = 3.89GGMTGLSAGDD1609 pKa = 3.56AVLGGDD1615 pKa = 4.74GNWALWGRR1623 pKa = 11.84VGGSKK1628 pKa = 10.76ASFTPSGVNGFDD1640 pKa = 3.4ADD1642 pKa = 3.37SWGVSLGLDD1651 pKa = 3.67GEE1653 pKa = 4.61VAPALRR1659 pKa = 11.84LGFGGFYY1666 pKa = 10.04IASDD1670 pKa = 3.82VEE1672 pKa = 4.49EE1673 pKa = 4.65NGAGANSTNDD1683 pKa = 2.81IAGYY1687 pKa = 10.9GFTTYY1692 pKa = 10.15MSWRR1696 pKa = 11.84PGAWYY1701 pKa = 10.9VNGALGYY1708 pKa = 11.3GMNTYY1713 pKa = 10.96DD1714 pKa = 4.89SRR1716 pKa = 11.84RR1717 pKa = 11.84TSLGSVNTADD1727 pKa = 3.56YY1728 pKa = 11.38DD1729 pKa = 3.72GTQFVARR1736 pKa = 11.84AEE1738 pKa = 4.03AGYY1741 pKa = 9.13MIVSGQWDD1749 pKa = 3.78LTPNVGLRR1757 pKa = 11.84YY1758 pKa = 10.03NRR1760 pKa = 11.84VDD1762 pKa = 2.89IDD1764 pKa = 4.21GYY1766 pKa = 11.54AEE1768 pKa = 4.27TGPLPISVNSQTVDD1782 pKa = 2.8SLRR1785 pKa = 11.84AVAGVNARR1793 pKa = 11.84YY1794 pKa = 7.84TMPLDD1799 pKa = 3.96GGGKK1803 pKa = 10.15LIPEE1807 pKa = 4.88FGVKK1811 pKa = 10.33LLGEE1815 pKa = 4.28LADD1818 pKa = 4.25PDD1820 pKa = 4.16GAITGQIVGGGVFVTQTTPRR1840 pKa = 11.84DD1841 pKa = 4.18DD1842 pKa = 3.04ISYY1845 pKa = 10.78GVGTGITYY1853 pKa = 7.98EE1854 pKa = 4.2ASDD1857 pKa = 3.62KK1858 pKa = 10.85FSIRR1862 pKa = 11.84VTYY1865 pKa = 10.53DD1866 pKa = 3.04GEE1868 pKa = 4.22FQSDD1872 pKa = 3.63YY1873 pKa = 11.63DD1874 pKa = 4.05EE1875 pKa = 4.6QALSAAIRR1883 pKa = 11.84WAFF1886 pKa = 3.13
MM1 pKa = 6.53TTRR4 pKa = 11.84HH5 pKa = 6.0SKK7 pKa = 10.87NITGTTGAIDD17 pKa = 3.58SSLLGSAYY25 pKa = 10.15SYY27 pKa = 11.25YY28 pKa = 10.94GGVVQTAHH36 pKa = 6.75SSGKK40 pKa = 7.27TASGVKK46 pKa = 9.76RR47 pKa = 11.84VFAAAMLSGASVLAVGIAGTGNAAAGTCVGPINEE81 pKa = 4.91TYY83 pKa = 9.52TCAGTFDD90 pKa = 3.9DD91 pKa = 4.81TIEE94 pKa = 4.24YY95 pKa = 10.57DD96 pKa = 3.56SGVDD100 pKa = 4.48DD101 pKa = 3.77VTLVLGPGSFIDD113 pKa = 3.62TTEE116 pKa = 3.86NTSEE120 pKa = 4.43HH121 pKa = 6.49DD122 pKa = 3.46HH123 pKa = 7.11DD124 pKa = 3.99NAGIVVVGEE133 pKa = 3.82SDD135 pKa = 3.34MAIVNNGSIFTGDD148 pKa = 2.73NTTWVEE154 pKa = 3.59TDD156 pKa = 3.15EE157 pKa = 4.59EE158 pKa = 4.44YY159 pKa = 10.99GYY161 pKa = 10.88YY162 pKa = 10.42AYY164 pKa = 10.88LGDD167 pKa = 3.89GHH169 pKa = 8.13HH170 pKa = 7.29GIAAYY175 pKa = 10.68SNWDD179 pKa = 3.45DD180 pKa = 3.74AAVEE184 pKa = 4.06NTAFGTIVTTSSEE197 pKa = 4.21SHH199 pKa = 5.85GMVAEE204 pKa = 4.1ALEE207 pKa = 4.94GSASADD213 pKa = 3.29NAGLIGTSGEE223 pKa = 4.13DD224 pKa = 3.1SHH226 pKa = 7.73GISATAKK233 pKa = 8.98YY234 pKa = 10.52SVDD237 pKa = 3.37VVNSGTITTDD247 pKa = 2.61GTYY250 pKa = 10.75SHH252 pKa = 7.26GIWAALDD259 pKa = 3.4SEE261 pKa = 4.52YY262 pKa = 10.93GYY264 pKa = 11.47GSFGGIIEE272 pKa = 4.34ISNSGSIEE280 pKa = 4.02TSGEE284 pKa = 4.13SASGISANAEE294 pKa = 3.89EE295 pKa = 5.1GDD297 pKa = 3.93VEE299 pKa = 4.31IANSGSIVTEE309 pKa = 3.78GDD311 pKa = 3.05EE312 pKa = 4.93ALGIWAHH319 pKa = 6.44GDD321 pKa = 3.43DD322 pKa = 3.74VDD324 pKa = 5.16VVNTADD330 pKa = 3.89GSIVTYY336 pKa = 10.26GADD339 pKa = 2.93AHH341 pKa = 6.06GVYY344 pKa = 10.3IFGDD348 pKa = 3.87TVSLSNAGTIEE359 pKa = 4.23TYY361 pKa = 10.98GEE363 pKa = 3.92DD364 pKa = 3.02AHH366 pKa = 8.29AVVAYY371 pKa = 10.3SGGIDD376 pKa = 3.14TTTIVNTGLIQATGEE391 pKa = 3.82EE392 pKa = 4.17ADD394 pKa = 4.7AIRR397 pKa = 11.84ASGPTVRR404 pKa = 11.84ITNDD408 pKa = 3.01VIEE411 pKa = 4.51EE412 pKa = 4.12DD413 pKa = 4.35EE414 pKa = 4.32EE415 pKa = 4.57VVDD418 pKa = 3.78YY419 pKa = 11.18GVIRR423 pKa = 11.84SEE425 pKa = 4.34DD426 pKa = 3.2GAAIHH431 pKa = 5.6VTEE434 pKa = 5.22ADD436 pKa = 4.07DD437 pKa = 4.5ARR439 pKa = 11.84LYY441 pKa = 11.41NNGAIYY447 pKa = 10.58GNVSIEE453 pKa = 3.94TDD455 pKa = 3.2EE456 pKa = 4.04YY457 pKa = 11.09GYY459 pKa = 11.42AVNTGSISSDD469 pKa = 3.56RR470 pKa = 11.84KK471 pKa = 8.89WKK473 pKa = 10.26AALAIDD479 pKa = 4.14AEE481 pKa = 4.41EE482 pKa = 5.05GDD484 pKa = 4.02VVVVNDD490 pKa = 4.19GDD492 pKa = 3.89IEE494 pKa = 4.4ASGRR498 pKa = 11.84KK499 pKa = 9.17AKK501 pKa = 10.35GVEE504 pKa = 3.84LEE506 pKa = 4.76GYY508 pKa = 10.32DD509 pKa = 3.51VTLTNTGSIEE519 pKa = 4.0TWGSDD524 pKa = 2.74GHH526 pKa = 7.82AVDD529 pKa = 5.38LEE531 pKa = 4.05GDD533 pKa = 3.67EE534 pKa = 4.91EE535 pKa = 4.52YY536 pKa = 11.63GEE538 pKa = 4.31VTLNNSGLIQASGEE552 pKa = 3.9EE553 pKa = 4.04ADD555 pKa = 3.81AVRR558 pKa = 11.84ASGEE562 pKa = 4.23TVRR565 pKa = 11.84ITNQAVLVPGEE576 pKa = 3.91TDD578 pKa = 3.64AYY580 pKa = 9.93DD581 pKa = 3.35TTIYY585 pKa = 10.69GIIRR589 pKa = 11.84SEE591 pKa = 4.17DD592 pKa = 3.16GAAIRR597 pKa = 11.84VDD599 pKa = 3.64EE600 pKa = 4.44SDD602 pKa = 3.18VVYY605 pKa = 10.79VVNDD609 pKa = 3.17GFIYY613 pKa = 10.72SNISIEE619 pKa = 4.33ADD621 pKa = 3.19DD622 pKa = 3.81YY623 pKa = 11.83AYY625 pKa = 11.1VSNAGSVSSDD635 pKa = 3.44RR636 pKa = 11.84RR637 pKa = 11.84WKK639 pKa = 10.41AAVSADD645 pKa = 3.67VEE647 pKa = 4.39EE648 pKa = 4.96GNAVIVNDD656 pKa = 4.22GEE658 pKa = 4.49IVTTASRR665 pKa = 11.84AAGIEE670 pKa = 3.9VDD672 pKa = 4.03VEE674 pKa = 4.07LGDD677 pKa = 4.07AYY679 pKa = 11.11VVNSGTITTGSLEE692 pKa = 4.04EE693 pKa = 4.61GEE695 pKa = 4.81PEE697 pKa = 4.56GGWRR701 pKa = 11.84SHH703 pKa = 6.74GIDD706 pKa = 3.44VFAEE710 pKa = 4.03DD711 pKa = 3.99TVLVLNTGDD720 pKa = 3.33VTTYY724 pKa = 8.85GHH726 pKa = 6.64KK727 pKa = 10.74AKK729 pKa = 10.61GIRR732 pKa = 11.84IEE734 pKa = 4.3TQEE737 pKa = 3.74GTAAGVNSGTIDD749 pKa = 2.92TWGEE753 pKa = 3.86DD754 pKa = 3.42STGLIVTATSRR765 pKa = 11.84EE766 pKa = 4.23HH767 pKa = 6.32YY768 pKa = 10.29DD769 pKa = 3.27GYY771 pKa = 11.28SYY773 pKa = 11.57FDD775 pKa = 3.12ISGIALAGNIGSVEE789 pKa = 4.07TRR791 pKa = 11.84GDD793 pKa = 3.4DD794 pKa = 3.36AMGVVAVAEE803 pKa = 4.64GNFAAALNVLGGSISTSGDD822 pKa = 3.37DD823 pKa = 3.37AHH825 pKa = 7.63GLVAVSGFEE834 pKa = 4.15SVEE837 pKa = 3.99DD838 pKa = 3.65ALEE841 pKa = 4.2GSEE844 pKa = 3.97YY845 pKa = 10.98SEE847 pKa = 4.64GGEE850 pKa = 3.92GSYY853 pKa = 10.9AIAVNGLPPQLLAGFSGIGTEE874 pKa = 4.1MGSGVLGEE882 pKa = 4.29IADD885 pKa = 4.14VLAEE889 pKa = 4.15ALPEE893 pKa = 4.27GEE895 pKa = 4.55YY896 pKa = 11.11DD897 pKa = 3.58PADD900 pKa = 3.62FRR902 pKa = 11.84STIVTTGDD910 pKa = 3.18GAIGVLAVSSNGGAVAGNLYY930 pKa = 10.48GDD932 pKa = 3.81VVTGVLHH939 pKa = 6.16EE940 pKa = 4.66EE941 pKa = 5.26GYY943 pKa = 10.78SVGGDD948 pKa = 3.28HH949 pKa = 7.46AYY951 pKa = 10.22GVAAMSVDD959 pKa = 3.42EE960 pKa = 4.88GGAIAFNAYY969 pKa = 9.23NASIVTNGAYY979 pKa = 10.18AHH981 pKa = 6.65GLLAISEE988 pKa = 5.08DD989 pKa = 3.8GDD991 pKa = 3.56AMAMNKK997 pKa = 9.37YY998 pKa = 10.26ASSIVTNGLGAVGIRR1013 pKa = 11.84VWTDD1017 pKa = 3.27GEE1019 pKa = 4.07SDD1021 pKa = 3.85EE1022 pKa = 4.87YY1023 pKa = 11.61SSADD1027 pKa = 3.28ALVWNVGSSVTTYY1040 pKa = 10.72GQGAQGIVAFSEE1052 pKa = 4.17EE1053 pKa = 4.25GDD1055 pKa = 3.72VTVANAPMTIGEE1067 pKa = 4.55GEE1069 pKa = 4.33DD1070 pKa = 3.53EE1071 pKa = 4.97EE1072 pKa = 4.65ILAGLIEE1079 pKa = 4.29THH1081 pKa = 6.64GDD1083 pKa = 3.28EE1084 pKa = 5.88ADD1086 pKa = 4.67GIWASGEE1093 pKa = 4.06SDD1095 pKa = 3.04VDD1097 pKa = 4.07VYY1099 pKa = 11.82NSGSIITHH1107 pKa = 5.56GEE1109 pKa = 3.83GANGISASGDD1119 pKa = 3.3EE1120 pKa = 4.07VDD1122 pKa = 4.55IDD1124 pKa = 3.81NSGTIQTSGDD1134 pKa = 3.37EE1135 pKa = 4.16AFGVDD1140 pKa = 3.36MRR1142 pKa = 11.84GWSVSLDD1149 pKa = 3.4NSGTISTSGKK1159 pKa = 8.38YY1160 pKa = 9.2AHH1162 pKa = 6.92AVVGDD1167 pKa = 4.17SYY1169 pKa = 12.15SDD1171 pKa = 3.46LTTTVYY1177 pKa = 11.08NSGTIEE1183 pKa = 4.04ATGKK1187 pKa = 9.1YY1188 pKa = 9.47ANAVRR1193 pKa = 11.84VSGPTVHH1200 pKa = 5.76VTNTEE1205 pKa = 4.02DD1206 pKa = 3.71GVISSGDD1213 pKa = 3.28VAIYY1217 pKa = 10.62AFEE1220 pKa = 4.08TKK1222 pKa = 9.91YY1223 pKa = 11.42ASIANHH1229 pKa = 6.26GEE1231 pKa = 4.17VEE1233 pKa = 4.29GSIHH1237 pKa = 6.13VSAYY1241 pKa = 10.32DD1242 pKa = 3.73YY1243 pKa = 11.55EE1244 pKa = 5.65DD1245 pKa = 3.57FASTYY1250 pKa = 9.94IVNTGSVSGDD1260 pKa = 2.56IDD1262 pKa = 3.76TSFGEE1267 pKa = 4.08SDD1269 pKa = 4.95DD1270 pKa = 4.46IIIVDD1275 pKa = 4.08GGTVGGAIFTGDD1287 pKa = 3.59GEE1289 pKa = 4.79DD1290 pKa = 3.49EE1291 pKa = 4.3VTISGTGVSIAKK1303 pKa = 9.79GIHH1306 pKa = 6.21GDD1308 pKa = 3.88GEE1310 pKa = 4.65GDD1312 pKa = 3.4LHH1314 pKa = 8.15VLFEE1318 pKa = 4.31QDD1320 pKa = 3.25DD1321 pKa = 4.39TVTFSDD1327 pKa = 4.85GIEE1330 pKa = 4.63GYY1332 pKa = 10.5AISNAHH1338 pKa = 5.75FVGFSSGTTVFDD1350 pKa = 3.58GVNIHH1355 pKa = 5.67TEE1357 pKa = 3.72EE1358 pKa = 5.47GEE1360 pKa = 4.07ILVGEE1365 pKa = 4.59DD1366 pKa = 2.86ATLATTIEE1374 pKa = 4.14NAFAFADD1381 pKa = 3.87EE1382 pKa = 4.34TSVLGRR1388 pKa = 11.84LKK1390 pKa = 10.7VVAGSSFGFSGDD1402 pKa = 3.55VVFDD1406 pKa = 3.57EE1407 pKa = 5.38GGTFEE1412 pKa = 4.77TGLTGDD1418 pKa = 3.63GAGVVTGDD1426 pKa = 3.51TVRR1429 pKa = 11.84FASGSTIHH1437 pKa = 6.48VDD1439 pKa = 3.02AGAGFTEE1446 pKa = 4.79TVGSDD1451 pKa = 3.24FLIASSEE1458 pKa = 4.33SEE1460 pKa = 4.44SGVTDD1465 pKa = 4.24DD1466 pKa = 5.08GASVTDD1472 pKa = 3.58NLILFKK1478 pKa = 10.45FLKK1481 pKa = 10.28VMNGDD1486 pKa = 3.76VVSEE1490 pKa = 4.41GAADD1494 pKa = 3.78EE1495 pKa = 4.3LFLRR1499 pKa = 11.84IEE1501 pKa = 4.19VEE1503 pKa = 3.93EE1504 pKa = 4.18TAFDD1508 pKa = 4.78LEE1510 pKa = 4.63TEE1512 pKa = 4.3AKK1514 pKa = 9.14GTGNLLSVASALDD1527 pKa = 3.82VYY1529 pKa = 11.07IEE1531 pKa = 4.21TQPLDD1536 pKa = 3.78NPLVQYY1542 pKa = 10.64LLQFEE1547 pKa = 4.66TEE1549 pKa = 4.26EE1550 pKa = 4.07EE1551 pKa = 3.88QLAALLKK1558 pKa = 10.11VIKK1561 pKa = 9.66DD1562 pKa = 3.85TVPDD1566 pKa = 3.87EE1567 pKa = 4.83SNASAGATIASTDD1580 pKa = 4.46LIYY1583 pKa = 11.38DD1584 pKa = 4.05MIMDD1588 pKa = 4.84RR1589 pKa = 11.84LSGGGFVVADD1599 pKa = 3.89GGMTGLSAGDD1609 pKa = 3.56AVLGGDD1615 pKa = 4.74GNWALWGRR1623 pKa = 11.84VGGSKK1628 pKa = 10.76ASFTPSGVNGFDD1640 pKa = 3.4ADD1642 pKa = 3.37SWGVSLGLDD1651 pKa = 3.67GEE1653 pKa = 4.61VAPALRR1659 pKa = 11.84LGFGGFYY1666 pKa = 10.04IASDD1670 pKa = 3.82VEE1672 pKa = 4.49EE1673 pKa = 4.65NGAGANSTNDD1683 pKa = 2.81IAGYY1687 pKa = 10.9GFTTYY1692 pKa = 10.15MSWRR1696 pKa = 11.84PGAWYY1701 pKa = 10.9VNGALGYY1708 pKa = 11.3GMNTYY1713 pKa = 10.96DD1714 pKa = 4.89SRR1716 pKa = 11.84RR1717 pKa = 11.84TSLGSVNTADD1727 pKa = 3.56YY1728 pKa = 11.38DD1729 pKa = 3.72GTQFVARR1736 pKa = 11.84AEE1738 pKa = 4.03AGYY1741 pKa = 9.13MIVSGQWDD1749 pKa = 3.78LTPNVGLRR1757 pKa = 11.84YY1758 pKa = 10.03NRR1760 pKa = 11.84VDD1762 pKa = 2.89IDD1764 pKa = 4.21GYY1766 pKa = 11.54AEE1768 pKa = 4.27TGPLPISVNSQTVDD1782 pKa = 2.8SLRR1785 pKa = 11.84AVAGVNARR1793 pKa = 11.84YY1794 pKa = 7.84TMPLDD1799 pKa = 3.96GGGKK1803 pKa = 10.15LIPEE1807 pKa = 4.88FGVKK1811 pKa = 10.33LLGEE1815 pKa = 4.28LADD1818 pKa = 4.25PDD1820 pKa = 4.16GAITGQIVGGGVFVTQTTPRR1840 pKa = 11.84DD1841 pKa = 4.18DD1842 pKa = 3.04ISYY1845 pKa = 10.78GVGTGITYY1853 pKa = 7.98EE1854 pKa = 4.2ASDD1857 pKa = 3.62KK1858 pKa = 10.85FSIRR1862 pKa = 11.84VTYY1865 pKa = 10.53DD1866 pKa = 3.04GEE1868 pKa = 4.22FQSDD1872 pKa = 3.63YY1873 pKa = 11.63DD1874 pKa = 4.05EE1875 pKa = 4.6QALSAAIRR1883 pKa = 11.84WAFF1886 pKa = 3.13
Molecular weight: 193.79 kDa
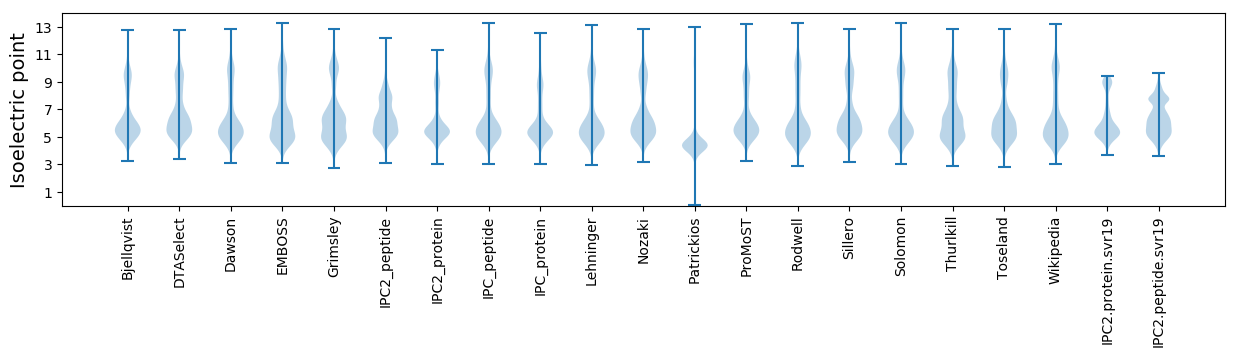
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A7HPY6|A7HPY6_PARL1 Copper-resistance protein CopA family OS=Parvibaculum lavamentivorans (strain DS-1 / DSM 13023 / NCIMB 13966) OX=402881 GN=Plav_0346 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.58GFRR19 pKa = 11.84ARR21 pKa = 11.84TQTVGGRR28 pKa = 11.84KK29 pKa = 8.9VLAARR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.53GRR39 pKa = 11.84KK40 pKa = 8.77RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.58GFRR19 pKa = 11.84ARR21 pKa = 11.84TQTVGGRR28 pKa = 11.84KK29 pKa = 8.9VLAARR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.53GRR39 pKa = 11.84KK40 pKa = 8.77RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1152854 |
29 |
3325 |
322.0 |
35.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.31 ± 0.058 | 0.811 ± 0.012 |
5.651 ± 0.031 | 6.518 ± 0.038 |
3.815 ± 0.027 | 8.689 ± 0.039 |
2.057 ± 0.018 | 5.15 ± 0.032 |
3.625 ± 0.035 | 9.87 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.594 ± 0.019 | 2.636 ± 0.023 |
5.112 ± 0.03 | 2.808 ± 0.022 |
7.026 ± 0.041 | 5.249 ± 0.032 |
5.167 ± 0.038 | 7.189 ± 0.032 |
1.353 ± 0.018 | 2.372 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
