
Hubei tombus-like virus 20
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.04
Get precalculated fractions of proteins
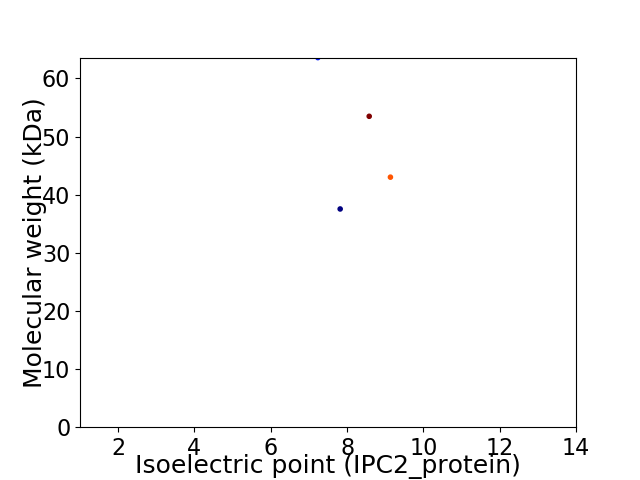
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGK2|A0A1L3KGK2_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 20 OX=1923267 PE=4 SV=1
MM1 pKa = 7.47VKK3 pKa = 10.38DD4 pKa = 4.26CLPTTDD10 pKa = 4.66CGAHH14 pKa = 5.06YY15 pKa = 9.62VQEE18 pKa = 4.4VVAPSGATSCDD29 pKa = 3.47GPPDD33 pKa = 4.24GSDD36 pKa = 3.2QSSSPAKK43 pKa = 10.04TRR45 pKa = 11.84DD46 pKa = 3.18QVIIPRR52 pKa = 11.84PSWIDD57 pKa = 3.39GTWDD61 pKa = 3.2VVTFLTPYY69 pKa = 9.85YY70 pKa = 8.29VQQVVSIAFPSEE82 pKa = 3.4ISATTVRR89 pKa = 11.84PYY91 pKa = 11.18ARR93 pKa = 11.84WVINSINTQFWWDD106 pKa = 3.58TQLPQWINPAEE117 pKa = 3.91HH118 pKa = 6.8VIYY121 pKa = 10.42NDD123 pKa = 2.84QGEE126 pKa = 4.5IVLVPFPFAVGFHH139 pKa = 6.72CPRR142 pKa = 11.84LLRR145 pKa = 11.84PLFEE149 pKa = 4.77GLGPEE154 pKa = 4.06PRR156 pKa = 11.84ILTEE160 pKa = 3.76TRR162 pKa = 11.84EE163 pKa = 3.79MRR165 pKa = 11.84RR166 pKa = 11.84WGRR169 pKa = 11.84YY170 pKa = 5.53ITITPVANATEE181 pKa = 4.48FKK183 pKa = 10.88GRR185 pKa = 11.84IAAASLQFNSALNPVHH201 pKa = 7.17AMVEE205 pKa = 4.36GGPDD209 pKa = 3.73PGPGPVDD216 pKa = 3.41PTPPPTLEE224 pKa = 3.95WARR227 pKa = 11.84FWCDD231 pKa = 2.81NYY233 pKa = 11.4SGTNPYY239 pKa = 10.55LYY241 pKa = 10.05TRR243 pKa = 11.84AGLTRR248 pKa = 11.84TWWSSSWLFEE258 pKa = 5.02DD259 pKa = 4.56NLSFDD264 pKa = 4.57LFNLNGNIIIPGNTPFQTSIVVLTSGGAPPALSFHH299 pKa = 4.75VTFRR303 pKa = 11.84VGNNVVFQGDD313 pKa = 4.12FSPGVAIPLFKK324 pKa = 10.43GRR326 pKa = 11.84RR327 pKa = 11.84AIRR330 pKa = 11.84STPIDD335 pKa = 3.45TDD337 pKa = 3.15EE338 pKa = 4.25LARR341 pKa = 11.84YY342 pKa = 6.94TITPEE347 pKa = 3.92FSFEE351 pKa = 4.08ALLQADD357 pKa = 4.13EE358 pKa = 4.85KK359 pKa = 11.16SYY361 pKa = 10.7SNNWIEE367 pKa = 4.43GAHH370 pKa = 6.4SRR372 pKa = 11.84QAWGKK377 pKa = 9.82GYY379 pKa = 11.04LEE381 pKa = 4.03FTPTRR386 pKa = 11.84EE387 pKa = 3.61WRR389 pKa = 11.84PLIRR393 pKa = 11.84APAGGRR399 pKa = 11.84VVIDD403 pKa = 3.24QASIKK408 pKa = 10.14RR409 pKa = 11.84DD410 pKa = 3.34IVDD413 pKa = 5.24LSGKK417 pKa = 7.38WHH419 pKa = 5.34VTYY422 pKa = 10.86ASNIDD427 pKa = 3.58PKK429 pKa = 11.13ASYY432 pKa = 10.25TMMYY436 pKa = 7.56GTHH439 pKa = 5.76YY440 pKa = 10.17QLCNEE445 pKa = 4.3TDD447 pKa = 3.38ASFQLFKK454 pKa = 11.06KK455 pKa = 9.7PVPARR460 pKa = 11.84DD461 pKa = 3.54DD462 pKa = 3.9GAIEE466 pKa = 3.98MALTLQTALPHH477 pKa = 5.76TFPAKK482 pKa = 10.54FNDD485 pKa = 3.92GGILPMILSQVKK497 pKa = 9.26RR498 pKa = 11.84VGSAGLAGLAKK509 pKa = 10.42GIVGSLMGGLQTIGTADD526 pKa = 3.47RR527 pKa = 11.84VIGYY531 pKa = 8.8PSMSTTYY538 pKa = 10.93GNNPVSLYY546 pKa = 10.78GGNGKK551 pKa = 8.33TNGRR555 pKa = 11.84RR556 pKa = 11.84GRR558 pKa = 11.84KK559 pKa = 8.84KK560 pKa = 10.01RR561 pKa = 11.84ANGRR565 pKa = 11.84DD566 pKa = 3.55AIASLMSRR574 pKa = 11.84TSNII578 pKa = 3.07
MM1 pKa = 7.47VKK3 pKa = 10.38DD4 pKa = 4.26CLPTTDD10 pKa = 4.66CGAHH14 pKa = 5.06YY15 pKa = 9.62VQEE18 pKa = 4.4VVAPSGATSCDD29 pKa = 3.47GPPDD33 pKa = 4.24GSDD36 pKa = 3.2QSSSPAKK43 pKa = 10.04TRR45 pKa = 11.84DD46 pKa = 3.18QVIIPRR52 pKa = 11.84PSWIDD57 pKa = 3.39GTWDD61 pKa = 3.2VVTFLTPYY69 pKa = 9.85YY70 pKa = 8.29VQQVVSIAFPSEE82 pKa = 3.4ISATTVRR89 pKa = 11.84PYY91 pKa = 11.18ARR93 pKa = 11.84WVINSINTQFWWDD106 pKa = 3.58TQLPQWINPAEE117 pKa = 3.91HH118 pKa = 6.8VIYY121 pKa = 10.42NDD123 pKa = 2.84QGEE126 pKa = 4.5IVLVPFPFAVGFHH139 pKa = 6.72CPRR142 pKa = 11.84LLRR145 pKa = 11.84PLFEE149 pKa = 4.77GLGPEE154 pKa = 4.06PRR156 pKa = 11.84ILTEE160 pKa = 3.76TRR162 pKa = 11.84EE163 pKa = 3.79MRR165 pKa = 11.84RR166 pKa = 11.84WGRR169 pKa = 11.84YY170 pKa = 5.53ITITPVANATEE181 pKa = 4.48FKK183 pKa = 10.88GRR185 pKa = 11.84IAAASLQFNSALNPVHH201 pKa = 7.17AMVEE205 pKa = 4.36GGPDD209 pKa = 3.73PGPGPVDD216 pKa = 3.41PTPPPTLEE224 pKa = 3.95WARR227 pKa = 11.84FWCDD231 pKa = 2.81NYY233 pKa = 11.4SGTNPYY239 pKa = 10.55LYY241 pKa = 10.05TRR243 pKa = 11.84AGLTRR248 pKa = 11.84TWWSSSWLFEE258 pKa = 5.02DD259 pKa = 4.56NLSFDD264 pKa = 4.57LFNLNGNIIIPGNTPFQTSIVVLTSGGAPPALSFHH299 pKa = 4.75VTFRR303 pKa = 11.84VGNNVVFQGDD313 pKa = 4.12FSPGVAIPLFKK324 pKa = 10.43GRR326 pKa = 11.84RR327 pKa = 11.84AIRR330 pKa = 11.84STPIDD335 pKa = 3.45TDD337 pKa = 3.15EE338 pKa = 4.25LARR341 pKa = 11.84YY342 pKa = 6.94TITPEE347 pKa = 3.92FSFEE351 pKa = 4.08ALLQADD357 pKa = 4.13EE358 pKa = 4.85KK359 pKa = 11.16SYY361 pKa = 10.7SNNWIEE367 pKa = 4.43GAHH370 pKa = 6.4SRR372 pKa = 11.84QAWGKK377 pKa = 9.82GYY379 pKa = 11.04LEE381 pKa = 4.03FTPTRR386 pKa = 11.84EE387 pKa = 3.61WRR389 pKa = 11.84PLIRR393 pKa = 11.84APAGGRR399 pKa = 11.84VVIDD403 pKa = 3.24QASIKK408 pKa = 10.14RR409 pKa = 11.84DD410 pKa = 3.34IVDD413 pKa = 5.24LSGKK417 pKa = 7.38WHH419 pKa = 5.34VTYY422 pKa = 10.86ASNIDD427 pKa = 3.58PKK429 pKa = 11.13ASYY432 pKa = 10.25TMMYY436 pKa = 7.56GTHH439 pKa = 5.76YY440 pKa = 10.17QLCNEE445 pKa = 4.3TDD447 pKa = 3.38ASFQLFKK454 pKa = 11.06KK455 pKa = 9.7PVPARR460 pKa = 11.84DD461 pKa = 3.54DD462 pKa = 3.9GAIEE466 pKa = 3.98MALTLQTALPHH477 pKa = 5.76TFPAKK482 pKa = 10.54FNDD485 pKa = 3.92GGILPMILSQVKK497 pKa = 9.26RR498 pKa = 11.84VGSAGLAGLAKK509 pKa = 10.42GIVGSLMGGLQTIGTADD526 pKa = 3.47RR527 pKa = 11.84VIGYY531 pKa = 8.8PSMSTTYY538 pKa = 10.93GNNPVSLYY546 pKa = 10.78GGNGKK551 pKa = 8.33TNGRR555 pKa = 11.84RR556 pKa = 11.84GRR558 pKa = 11.84KK559 pKa = 8.84KK560 pKa = 10.01RR561 pKa = 11.84ANGRR565 pKa = 11.84DD566 pKa = 3.55AIASLMSRR574 pKa = 11.84TSNII578 pKa = 3.07
Molecular weight: 63.59 kDa
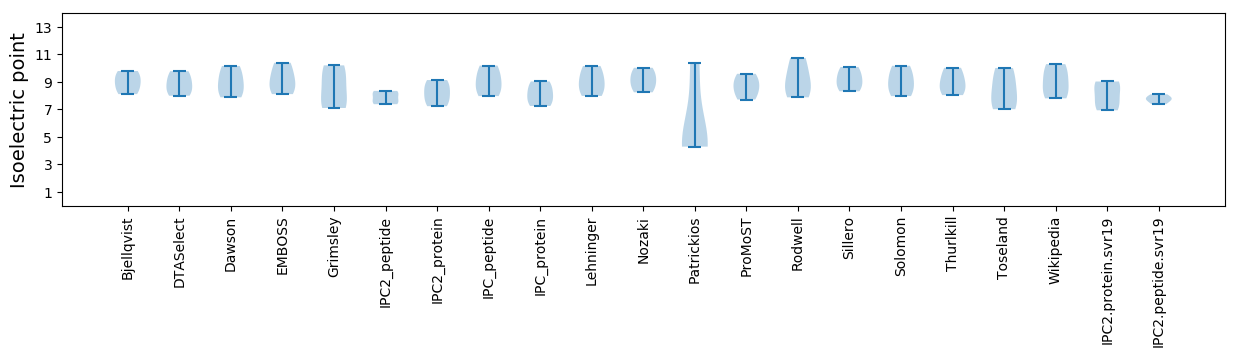
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGK2|A0A1L3KGK2_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 20 OX=1923267 PE=4 SV=1
MM1 pKa = 7.71NDD3 pKa = 3.03RR4 pKa = 11.84GNRR7 pKa = 11.84VPLGGKK13 pKa = 8.69RR14 pKa = 11.84PKK16 pKa = 8.23MTLAQVVAKK25 pKa = 8.16FAPYY29 pKa = 10.51LKK31 pKa = 10.17PRR33 pKa = 11.84VDD35 pKa = 2.99ALQRR39 pKa = 11.84TEE41 pKa = 4.19RR42 pKa = 11.84LYY44 pKa = 11.23KK45 pKa = 10.34EE46 pKa = 5.24LMAKK50 pKa = 8.49TPPHH54 pKa = 5.93SAVKK58 pKa = 9.15KK59 pKa = 9.53QRR61 pKa = 11.84VATRR65 pKa = 11.84PNKK68 pKa = 10.37CYY70 pKa = 10.37EE71 pKa = 4.56CGSKK75 pKa = 7.33QHH77 pKa = 5.87RR78 pKa = 11.84TYY80 pKa = 10.54GTDD83 pKa = 3.1RR84 pKa = 11.84EE85 pKa = 4.43GWVACEE91 pKa = 4.38CFRR94 pKa = 11.84CGTEE98 pKa = 4.18YY99 pKa = 10.42FYY101 pKa = 11.19HH102 pKa = 7.15PNQGVQSSAGYY113 pKa = 7.41TAIGTKK119 pKa = 10.13PVVNNSASWAGVVKK133 pKa = 10.61SNKK136 pKa = 9.4RR137 pKa = 11.84PQTPSSDD144 pKa = 2.97WKK146 pKa = 10.95KK147 pKa = 10.87DD148 pKa = 3.2WNKK151 pKa = 10.05VKK153 pKa = 10.84DD154 pKa = 3.76KK155 pKa = 11.16VSNQTTTATPQAPQSPQPPKK175 pKa = 10.45LSRR178 pKa = 11.84AQRR181 pKa = 11.84AKK183 pKa = 10.17RR184 pKa = 11.84AKK186 pKa = 9.25EE187 pKa = 3.54LRR189 pKa = 11.84AEE191 pKa = 4.2KK192 pKa = 10.1LAKK195 pKa = 8.62QAKK198 pKa = 9.6PITQLKK204 pKa = 9.95SSSEE208 pKa = 4.32SPDD211 pKa = 3.04TALDD215 pKa = 3.44VTEE218 pKa = 4.46IKK220 pKa = 10.0EE221 pKa = 4.04DD222 pKa = 3.57KK223 pKa = 9.65PIKK226 pKa = 10.46VNGEE230 pKa = 3.97IITAEE235 pKa = 4.07ALTASINQVLKK246 pKa = 10.01TVDD249 pKa = 3.46VSDD252 pKa = 5.12DD253 pKa = 3.65LPTYY257 pKa = 8.91EE258 pKa = 5.37QYY260 pKa = 11.39LKK262 pKa = 10.9DD263 pKa = 4.21FRR265 pKa = 11.84TEE267 pKa = 3.67SHH269 pKa = 6.04KK270 pKa = 11.05QPGEE274 pKa = 3.7VGKK277 pKa = 9.34WIKK280 pKa = 10.45SAEE283 pKa = 4.29QILQSWPTRR292 pKa = 11.84LLVCTPTGVYY302 pKa = 9.15RR303 pKa = 11.84VKK305 pKa = 10.69RR306 pKa = 11.84DD307 pKa = 3.51LEE309 pKa = 4.25STKK312 pKa = 10.43PYY314 pKa = 10.39LLSYY318 pKa = 10.9LKK320 pKa = 10.55LLKK323 pKa = 10.63AADD326 pKa = 4.55LEE328 pKa = 5.1DD329 pKa = 5.62LDD331 pKa = 5.31LSNWDD336 pKa = 3.67QQISAAAMMVATKK349 pKa = 9.72PLRR352 pKa = 11.84ASWRR356 pKa = 11.84RR357 pKa = 11.84AKK359 pKa = 10.3GRR361 pKa = 11.84RR362 pKa = 11.84KK363 pKa = 8.56TGGTNPPTRR372 pKa = 11.84EE373 pKa = 4.04CKK375 pKa = 10.02EE376 pKa = 4.02SSKK379 pKa = 11.18EE380 pKa = 3.81MAGTT384 pKa = 3.52
MM1 pKa = 7.71NDD3 pKa = 3.03RR4 pKa = 11.84GNRR7 pKa = 11.84VPLGGKK13 pKa = 8.69RR14 pKa = 11.84PKK16 pKa = 8.23MTLAQVVAKK25 pKa = 8.16FAPYY29 pKa = 10.51LKK31 pKa = 10.17PRR33 pKa = 11.84VDD35 pKa = 2.99ALQRR39 pKa = 11.84TEE41 pKa = 4.19RR42 pKa = 11.84LYY44 pKa = 11.23KK45 pKa = 10.34EE46 pKa = 5.24LMAKK50 pKa = 8.49TPPHH54 pKa = 5.93SAVKK58 pKa = 9.15KK59 pKa = 9.53QRR61 pKa = 11.84VATRR65 pKa = 11.84PNKK68 pKa = 10.37CYY70 pKa = 10.37EE71 pKa = 4.56CGSKK75 pKa = 7.33QHH77 pKa = 5.87RR78 pKa = 11.84TYY80 pKa = 10.54GTDD83 pKa = 3.1RR84 pKa = 11.84EE85 pKa = 4.43GWVACEE91 pKa = 4.38CFRR94 pKa = 11.84CGTEE98 pKa = 4.18YY99 pKa = 10.42FYY101 pKa = 11.19HH102 pKa = 7.15PNQGVQSSAGYY113 pKa = 7.41TAIGTKK119 pKa = 10.13PVVNNSASWAGVVKK133 pKa = 10.61SNKK136 pKa = 9.4RR137 pKa = 11.84PQTPSSDD144 pKa = 2.97WKK146 pKa = 10.95KK147 pKa = 10.87DD148 pKa = 3.2WNKK151 pKa = 10.05VKK153 pKa = 10.84DD154 pKa = 3.76KK155 pKa = 11.16VSNQTTTATPQAPQSPQPPKK175 pKa = 10.45LSRR178 pKa = 11.84AQRR181 pKa = 11.84AKK183 pKa = 10.17RR184 pKa = 11.84AKK186 pKa = 9.25EE187 pKa = 3.54LRR189 pKa = 11.84AEE191 pKa = 4.2KK192 pKa = 10.1LAKK195 pKa = 8.62QAKK198 pKa = 9.6PITQLKK204 pKa = 9.95SSSEE208 pKa = 4.32SPDD211 pKa = 3.04TALDD215 pKa = 3.44VTEE218 pKa = 4.46IKK220 pKa = 10.0EE221 pKa = 4.04DD222 pKa = 3.57KK223 pKa = 9.65PIKK226 pKa = 10.46VNGEE230 pKa = 3.97IITAEE235 pKa = 4.07ALTASINQVLKK246 pKa = 10.01TVDD249 pKa = 3.46VSDD252 pKa = 5.12DD253 pKa = 3.65LPTYY257 pKa = 8.91EE258 pKa = 5.37QYY260 pKa = 11.39LKK262 pKa = 10.9DD263 pKa = 4.21FRR265 pKa = 11.84TEE267 pKa = 3.67SHH269 pKa = 6.04KK270 pKa = 11.05QPGEE274 pKa = 3.7VGKK277 pKa = 9.34WIKK280 pKa = 10.45SAEE283 pKa = 4.29QILQSWPTRR292 pKa = 11.84LLVCTPTGVYY302 pKa = 9.15RR303 pKa = 11.84VKK305 pKa = 10.69RR306 pKa = 11.84DD307 pKa = 3.51LEE309 pKa = 4.25STKK312 pKa = 10.43PYY314 pKa = 10.39LLSYY318 pKa = 10.9LKK320 pKa = 10.55LLKK323 pKa = 10.63AADD326 pKa = 4.55LEE328 pKa = 5.1DD329 pKa = 5.62LDD331 pKa = 5.31LSNWDD336 pKa = 3.67QQISAAAMMVATKK349 pKa = 9.72PLRR352 pKa = 11.84ASWRR356 pKa = 11.84RR357 pKa = 11.84AKK359 pKa = 10.3GRR361 pKa = 11.84RR362 pKa = 11.84KK363 pKa = 8.56TGGTNPPTRR372 pKa = 11.84EE373 pKa = 4.04CKK375 pKa = 10.02EE376 pKa = 4.02SSKK379 pKa = 11.18EE380 pKa = 3.81MAGTT384 pKa = 3.52
Molecular weight: 43.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1768 |
336 |
578 |
442.0 |
49.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.749 ± 0.685 | 1.867 ± 0.378 |
4.864 ± 0.124 | 4.921 ± 0.566 |
3.45 ± 0.647 | 7.24 ± 0.683 |
1.867 ± 0.296 | 5.09 ± 0.634 |
6.335 ± 1.341 | 7.862 ± 0.567 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.376 ± 0.393 | 4.186 ± 0.643 |
6.731 ± 0.619 | 3.846 ± 0.417 |
6.505 ± 0.181 | 6.787 ± 0.235 |
6.505 ± 0.771 | 6.165 ± 0.176 |
2.036 ± 0.337 | 3.62 ± 0.287 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
