
Inoviridae sp.
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; unclassified Inoviridae
Average proteome isoelectric point is 5.56
Get precalculated fractions of proteins
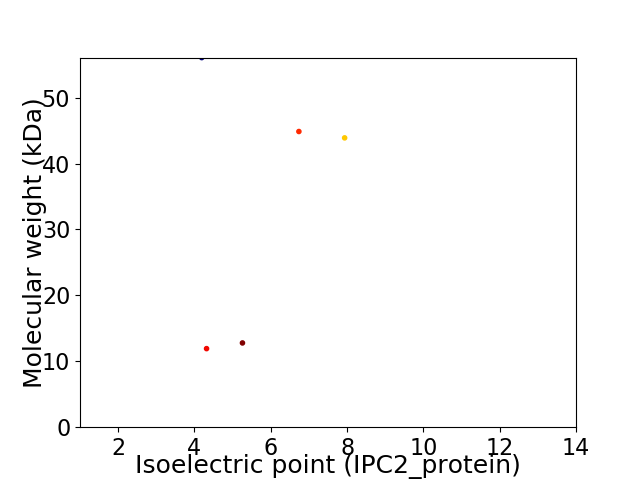
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345N1M7|A0A345N1M7_9VIRU Uncharacterized protein OS=Inoviridae sp. OX=2219103 PE=4 SV=1
MM1 pKa = 7.85KK2 pKa = 10.68YY3 pKa = 10.52LILPMALMSAFFVSANEE20 pKa = 3.95PDD22 pKa = 4.69LDD24 pKa = 4.3DD25 pKa = 4.87LVSPKK30 pKa = 9.98QVPEE34 pKa = 3.59IKK36 pKa = 10.6YY37 pKa = 9.09FNPNNSLLFNSPKK50 pKa = 10.47DD51 pKa = 3.54AFEE54 pKa = 4.28SRR56 pKa = 11.84FDD58 pKa = 3.53EE59 pKa = 4.51SQFCEE64 pKa = 4.72GTWSCTIIDD73 pKa = 3.75EE74 pKa = 4.71SEE76 pKa = 4.49TVTGTSVKK84 pKa = 10.36YY85 pKa = 7.86YY86 pKa = 10.33RR87 pKa = 11.84KK88 pKa = 9.46YY89 pKa = 9.36EE90 pKa = 4.15TYY92 pKa = 10.44SKK94 pKa = 10.48CGYY97 pKa = 8.94PDD99 pKa = 3.42YY100 pKa = 10.83YY101 pKa = 10.71KK102 pKa = 10.85CNRR105 pKa = 11.84KK106 pKa = 8.75EE107 pKa = 3.71RR108 pKa = 11.84GFAEE112 pKa = 4.45VYY114 pKa = 10.36PIVEE118 pKa = 4.6EE119 pKa = 4.26FTYY122 pKa = 10.54SCPPDD127 pKa = 3.52NRR129 pKa = 11.84PLHH132 pKa = 6.36KK133 pKa = 10.16IPVPINPVPTDD144 pKa = 3.72GPKK147 pKa = 10.45FMCAKK152 pKa = 10.12PLDD155 pKa = 4.21NEE157 pKa = 4.17PDD159 pKa = 3.68PNAKK163 pKa = 9.77CDD165 pKa = 3.68EE166 pKa = 4.53FGNNSMLPPKK176 pKa = 10.38AGVGSEE182 pKa = 4.18GQNACYY188 pKa = 9.48TNPSNGLSCQYY199 pKa = 10.68VQGGDD204 pKa = 3.03NFTATGKK211 pKa = 10.01QCTGDD216 pKa = 3.53EE217 pKa = 4.19NDD219 pKa = 3.6YY220 pKa = 11.4GDD222 pKa = 4.6RR223 pKa = 11.84PTPEE227 pKa = 4.78PPPDD231 pKa = 3.84GADD234 pKa = 3.48PNCYY238 pKa = 10.2NYY240 pKa = 10.7GSQGQVLICDD250 pKa = 3.52VDD252 pKa = 4.01PNEE255 pKa = 4.38GCNPLTINGTTQYY268 pKa = 10.4QCPSGCGSIDD278 pKa = 3.07GVYY281 pKa = 10.06FCSYY285 pKa = 10.64DD286 pKa = 3.92DD287 pKa = 4.48KK288 pKa = 11.66DD289 pKa = 4.03GDD291 pKa = 5.38GIPDD295 pKa = 3.84DD296 pKa = 4.56KK297 pKa = 11.42NGNGVPDD304 pKa = 3.83KK305 pKa = 11.53DD306 pKa = 3.77EE307 pKa = 4.14TCVNGKK313 pKa = 9.56CKK315 pKa = 10.39PNTPDD320 pKa = 3.38DD321 pKa = 3.99TTPPTTEE328 pKa = 4.29KK329 pKa = 10.89PDD331 pKa = 3.41MTEE334 pKa = 3.54TNTRR338 pKa = 11.84LDD340 pKa = 3.75GVIGEE345 pKa = 4.73LDD347 pKa = 3.81SIGGKK352 pKa = 9.24INKK355 pKa = 7.96TNQTLDD361 pKa = 4.31GINSGIQGIKK371 pKa = 9.82QGQDD375 pKa = 2.76KK376 pKa = 10.78TNGLLTGMSKK386 pKa = 9.22DD387 pKa = 3.17TGLIANNTDD396 pKa = 4.02AIAGNTKK403 pKa = 10.35GLLEE407 pKa = 4.52TVSKK411 pKa = 9.99TDD413 pKa = 3.04VGNTFNPDD421 pKa = 3.11LSTGFYY427 pKa = 9.98EE428 pKa = 4.55SSYY431 pKa = 10.87EE432 pKa = 4.08NGFEE436 pKa = 4.66GIWAEE441 pKa = 3.91KK442 pKa = 10.53SILFEE447 pKa = 3.91QTEE450 pKa = 4.79TIQFLQQFRR459 pKa = 11.84FNAGGSPPDD468 pKa = 3.6TQLCFNMGGSMDD480 pKa = 4.88FGCADD485 pKa = 4.83LPTPSPQLLAILKK498 pKa = 9.92IFILITAAFLCRR510 pKa = 11.84ALIFGGG516 pKa = 3.67
MM1 pKa = 7.85KK2 pKa = 10.68YY3 pKa = 10.52LILPMALMSAFFVSANEE20 pKa = 3.95PDD22 pKa = 4.69LDD24 pKa = 4.3DD25 pKa = 4.87LVSPKK30 pKa = 9.98QVPEE34 pKa = 3.59IKK36 pKa = 10.6YY37 pKa = 9.09FNPNNSLLFNSPKK50 pKa = 10.47DD51 pKa = 3.54AFEE54 pKa = 4.28SRR56 pKa = 11.84FDD58 pKa = 3.53EE59 pKa = 4.51SQFCEE64 pKa = 4.72GTWSCTIIDD73 pKa = 3.75EE74 pKa = 4.71SEE76 pKa = 4.49TVTGTSVKK84 pKa = 10.36YY85 pKa = 7.86YY86 pKa = 10.33RR87 pKa = 11.84KK88 pKa = 9.46YY89 pKa = 9.36EE90 pKa = 4.15TYY92 pKa = 10.44SKK94 pKa = 10.48CGYY97 pKa = 8.94PDD99 pKa = 3.42YY100 pKa = 10.83YY101 pKa = 10.71KK102 pKa = 10.85CNRR105 pKa = 11.84KK106 pKa = 8.75EE107 pKa = 3.71RR108 pKa = 11.84GFAEE112 pKa = 4.45VYY114 pKa = 10.36PIVEE118 pKa = 4.6EE119 pKa = 4.26FTYY122 pKa = 10.54SCPPDD127 pKa = 3.52NRR129 pKa = 11.84PLHH132 pKa = 6.36KK133 pKa = 10.16IPVPINPVPTDD144 pKa = 3.72GPKK147 pKa = 10.45FMCAKK152 pKa = 10.12PLDD155 pKa = 4.21NEE157 pKa = 4.17PDD159 pKa = 3.68PNAKK163 pKa = 9.77CDD165 pKa = 3.68EE166 pKa = 4.53FGNNSMLPPKK176 pKa = 10.38AGVGSEE182 pKa = 4.18GQNACYY188 pKa = 9.48TNPSNGLSCQYY199 pKa = 10.68VQGGDD204 pKa = 3.03NFTATGKK211 pKa = 10.01QCTGDD216 pKa = 3.53EE217 pKa = 4.19NDD219 pKa = 3.6YY220 pKa = 11.4GDD222 pKa = 4.6RR223 pKa = 11.84PTPEE227 pKa = 4.78PPPDD231 pKa = 3.84GADD234 pKa = 3.48PNCYY238 pKa = 10.2NYY240 pKa = 10.7GSQGQVLICDD250 pKa = 3.52VDD252 pKa = 4.01PNEE255 pKa = 4.38GCNPLTINGTTQYY268 pKa = 10.4QCPSGCGSIDD278 pKa = 3.07GVYY281 pKa = 10.06FCSYY285 pKa = 10.64DD286 pKa = 3.92DD287 pKa = 4.48KK288 pKa = 11.66DD289 pKa = 4.03GDD291 pKa = 5.38GIPDD295 pKa = 3.84DD296 pKa = 4.56KK297 pKa = 11.42NGNGVPDD304 pKa = 3.83KK305 pKa = 11.53DD306 pKa = 3.77EE307 pKa = 4.14TCVNGKK313 pKa = 9.56CKK315 pKa = 10.39PNTPDD320 pKa = 3.38DD321 pKa = 3.99TTPPTTEE328 pKa = 4.29KK329 pKa = 10.89PDD331 pKa = 3.41MTEE334 pKa = 3.54TNTRR338 pKa = 11.84LDD340 pKa = 3.75GVIGEE345 pKa = 4.73LDD347 pKa = 3.81SIGGKK352 pKa = 9.24INKK355 pKa = 7.96TNQTLDD361 pKa = 4.31GINSGIQGIKK371 pKa = 9.82QGQDD375 pKa = 2.76KK376 pKa = 10.78TNGLLTGMSKK386 pKa = 9.22DD387 pKa = 3.17TGLIANNTDD396 pKa = 4.02AIAGNTKK403 pKa = 10.35GLLEE407 pKa = 4.52TVSKK411 pKa = 9.99TDD413 pKa = 3.04VGNTFNPDD421 pKa = 3.11LSTGFYY427 pKa = 9.98EE428 pKa = 4.55SSYY431 pKa = 10.87EE432 pKa = 4.08NGFEE436 pKa = 4.66GIWAEE441 pKa = 3.91KK442 pKa = 10.53SILFEE447 pKa = 3.91QTEE450 pKa = 4.79TIQFLQQFRR459 pKa = 11.84FNAGGSPPDD468 pKa = 3.6TQLCFNMGGSMDD480 pKa = 4.88FGCADD485 pKa = 4.83LPTPSPQLLAILKK498 pKa = 9.92IFILITAAFLCRR510 pKa = 11.84ALIFGGG516 pKa = 3.67
Molecular weight: 56.07 kDa
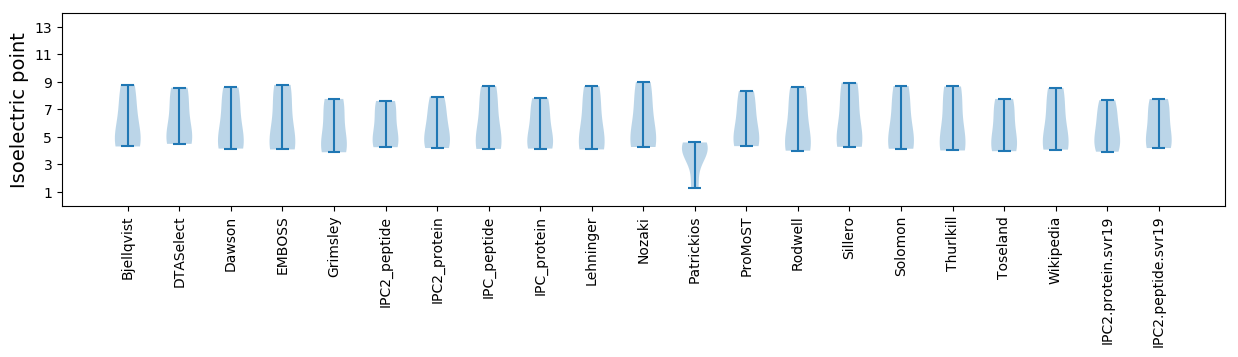
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345N1N0|A0A345N1N0_9VIRU DNA replication initiation protein OS=Inoviridae sp. OX=2219103 PE=4 SV=1
MM1 pKa = 7.49HH2 pKa = 7.05YY3 pKa = 9.95KK4 pKa = 10.72YY5 pKa = 10.97SDD7 pKa = 3.29TAQNLIDD14 pKa = 3.72NRR16 pKa = 11.84IKK18 pKa = 10.72RR19 pKa = 11.84FKK21 pKa = 10.43KK22 pKa = 9.82HH23 pKa = 5.69EE24 pKa = 4.45ANQTIIDD31 pKa = 3.99HH32 pKa = 6.84LSFSFPLSDD41 pKa = 4.43LRR43 pKa = 11.84HH44 pKa = 6.17CKK46 pKa = 9.86RR47 pKa = 11.84AGSIGSTVDD56 pKa = 3.12TQTLFPVVPNIKK68 pKa = 10.28EE69 pKa = 4.0EE70 pKa = 4.17FSTEE74 pKa = 3.62GLTPDD79 pKa = 3.75EE80 pKa = 4.25VLKK83 pKa = 11.15SLEE86 pKa = 4.19TQKK89 pKa = 10.58QRR91 pKa = 11.84INSRR95 pKa = 11.84MSDD98 pKa = 3.57FYY100 pKa = 11.14INTLRR105 pKa = 11.84VFSRR109 pKa = 11.84YY110 pKa = 8.51VLGFDD115 pKa = 4.99LSAPRR120 pKa = 11.84DD121 pKa = 3.77KK122 pKa = 11.3GFHH125 pKa = 6.58GYY127 pKa = 9.25HH128 pKa = 6.3NSMNLVTSQGNKK140 pKa = 9.48VGFVGIGGQRR150 pKa = 11.84DD151 pKa = 3.36TVYY154 pKa = 10.61FQISGEE160 pKa = 4.34GCKK163 pKa = 10.26HH164 pKa = 5.88LWSHH168 pKa = 5.58TTPFILHH175 pKa = 6.21HH176 pKa = 6.53WLSKK180 pKa = 10.47VLSISNLSRR189 pKa = 11.84IDD191 pKa = 3.49IARR194 pKa = 11.84DD195 pKa = 3.41CYY197 pKa = 11.5DD198 pKa = 4.37DD199 pKa = 5.05VFNCVNAEE207 pKa = 3.64KK208 pKa = 10.99DD209 pKa = 3.64FFNSAFRR216 pKa = 11.84RR217 pKa = 11.84NKK219 pKa = 10.16GGPSPKK225 pKa = 9.44MGNHH229 pKa = 6.81DD230 pKa = 4.64SISIDD235 pKa = 2.95RR236 pKa = 11.84VFDD239 pKa = 3.61VEE241 pKa = 4.26MKK243 pKa = 10.79SFGKK247 pKa = 8.75RR248 pKa = 11.84TSPVYY253 pKa = 9.54WRR255 pKa = 11.84VYY257 pKa = 9.8NKK259 pKa = 10.39KK260 pKa = 10.26LEE262 pKa = 3.94QGIDD266 pKa = 3.52DD267 pKa = 5.68DD268 pKa = 4.64SLVWYY273 pKa = 10.28RR274 pKa = 11.84NEE276 pKa = 3.87VEE278 pKa = 4.14LKK280 pKa = 9.76KK281 pKa = 10.19WSVDD285 pKa = 3.44CLLDD289 pKa = 4.07PDD291 pKa = 4.16SAFAGICAFSQQMINTDD308 pKa = 3.58GVQTSSSSKK317 pKa = 9.2VSKK320 pKa = 10.81AGTDD324 pKa = 3.2LASRR328 pKa = 11.84VKK330 pKa = 9.4WVRR333 pKa = 11.84RR334 pKa = 11.84MCGKK338 pKa = 10.32ALADD342 pKa = 3.07ILEE345 pKa = 4.33ITEE348 pKa = 4.74GDD350 pKa = 3.64MQTVLGLLVPHH361 pKa = 7.13KK362 pKa = 10.32YY363 pKa = 8.39VTGKK367 pKa = 10.51SLDD370 pKa = 3.26VPNIYY375 pKa = 10.52KK376 pKa = 10.5HH377 pKa = 6.45ILTEE381 pKa = 3.93QLRR384 pKa = 11.84SHH386 pKa = 7.02
MM1 pKa = 7.49HH2 pKa = 7.05YY3 pKa = 9.95KK4 pKa = 10.72YY5 pKa = 10.97SDD7 pKa = 3.29TAQNLIDD14 pKa = 3.72NRR16 pKa = 11.84IKK18 pKa = 10.72RR19 pKa = 11.84FKK21 pKa = 10.43KK22 pKa = 9.82HH23 pKa = 5.69EE24 pKa = 4.45ANQTIIDD31 pKa = 3.99HH32 pKa = 6.84LSFSFPLSDD41 pKa = 4.43LRR43 pKa = 11.84HH44 pKa = 6.17CKK46 pKa = 9.86RR47 pKa = 11.84AGSIGSTVDD56 pKa = 3.12TQTLFPVVPNIKK68 pKa = 10.28EE69 pKa = 4.0EE70 pKa = 4.17FSTEE74 pKa = 3.62GLTPDD79 pKa = 3.75EE80 pKa = 4.25VLKK83 pKa = 11.15SLEE86 pKa = 4.19TQKK89 pKa = 10.58QRR91 pKa = 11.84INSRR95 pKa = 11.84MSDD98 pKa = 3.57FYY100 pKa = 11.14INTLRR105 pKa = 11.84VFSRR109 pKa = 11.84YY110 pKa = 8.51VLGFDD115 pKa = 4.99LSAPRR120 pKa = 11.84DD121 pKa = 3.77KK122 pKa = 11.3GFHH125 pKa = 6.58GYY127 pKa = 9.25HH128 pKa = 6.3NSMNLVTSQGNKK140 pKa = 9.48VGFVGIGGQRR150 pKa = 11.84DD151 pKa = 3.36TVYY154 pKa = 10.61FQISGEE160 pKa = 4.34GCKK163 pKa = 10.26HH164 pKa = 5.88LWSHH168 pKa = 5.58TTPFILHH175 pKa = 6.21HH176 pKa = 6.53WLSKK180 pKa = 10.47VLSISNLSRR189 pKa = 11.84IDD191 pKa = 3.49IARR194 pKa = 11.84DD195 pKa = 3.41CYY197 pKa = 11.5DD198 pKa = 4.37DD199 pKa = 5.05VFNCVNAEE207 pKa = 3.64KK208 pKa = 10.99DD209 pKa = 3.64FFNSAFRR216 pKa = 11.84RR217 pKa = 11.84NKK219 pKa = 10.16GGPSPKK225 pKa = 9.44MGNHH229 pKa = 6.81DD230 pKa = 4.64SISIDD235 pKa = 2.95RR236 pKa = 11.84VFDD239 pKa = 3.61VEE241 pKa = 4.26MKK243 pKa = 10.79SFGKK247 pKa = 8.75RR248 pKa = 11.84TSPVYY253 pKa = 9.54WRR255 pKa = 11.84VYY257 pKa = 9.8NKK259 pKa = 10.39KK260 pKa = 10.26LEE262 pKa = 3.94QGIDD266 pKa = 3.52DD267 pKa = 5.68DD268 pKa = 4.64SLVWYY273 pKa = 10.28RR274 pKa = 11.84NEE276 pKa = 3.87VEE278 pKa = 4.14LKK280 pKa = 9.76KK281 pKa = 10.19WSVDD285 pKa = 3.44CLLDD289 pKa = 4.07PDD291 pKa = 4.16SAFAGICAFSQQMINTDD308 pKa = 3.58GVQTSSSSKK317 pKa = 9.2VSKK320 pKa = 10.81AGTDD324 pKa = 3.2LASRR328 pKa = 11.84VKK330 pKa = 9.4WVRR333 pKa = 11.84RR334 pKa = 11.84MCGKK338 pKa = 10.32ALADD342 pKa = 3.07ILEE345 pKa = 4.33ITEE348 pKa = 4.74GDD350 pKa = 3.64MQTVLGLLVPHH361 pKa = 7.13KK362 pKa = 10.32YY363 pKa = 8.39VTGKK367 pKa = 10.51SLDD370 pKa = 3.26VPNIYY375 pKa = 10.52KK376 pKa = 10.5HH377 pKa = 6.45ILTEE381 pKa = 3.93QLRR384 pKa = 11.84SHH386 pKa = 7.02
Molecular weight: 43.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1511 |
105 |
516 |
302.2 |
33.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.427 ± 1.273 | 2.184 ± 0.806 |
7.346 ± 0.549 | 5.361 ± 0.632 |
5.493 ± 0.397 | 7.28 ± 1.173 |
2.449 ± 0.984 | 6.353 ± 0.907 |
7.015 ± 0.847 | 7.081 ± 0.92 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.052 ± 0.41 | 5.493 ± 0.894 |
5.162 ± 1.355 | 4.236 ± 0.686 |
3.309 ± 0.811 | 6.949 ± 0.935 |
5.824 ± 0.937 | 5.956 ± 0.962 |
1.059 ± 0.397 | 3.971 ± 0.451 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
