
Aedes camptorhynchus negev-like virus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.74
Get precalculated fractions of proteins
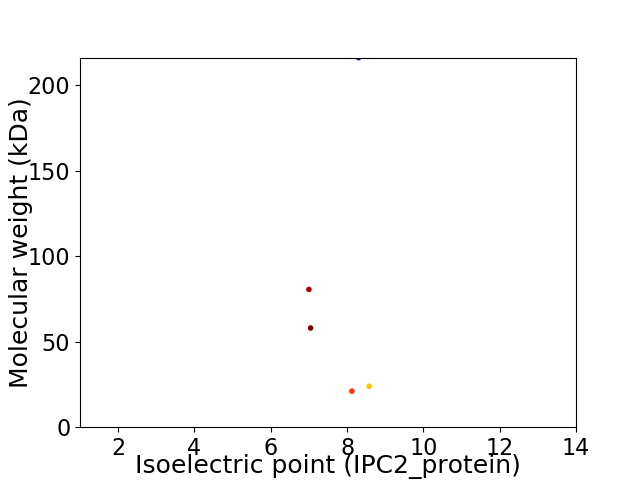
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z2RT39|A0A1Z2RT39_9VIRU RdRp OS=Aedes camptorhynchus negev-like virus OX=2010268 PE=4 SV=1
MM1 pKa = 8.01DD2 pKa = 5.02SSNVLQFFFDD12 pKa = 3.72TVLPGSSLEE21 pKa = 4.1FRR23 pKa = 11.84EE24 pKa = 4.03HH25 pKa = 5.67DD26 pKa = 3.37HH27 pKa = 7.47LRR29 pKa = 11.84FEE31 pKa = 5.07YY32 pKa = 7.11EE33 pKa = 3.99TPMKK37 pKa = 10.84AEE39 pKa = 4.63GIMFEE44 pKa = 4.55LSVPTFKK51 pKa = 10.91KK52 pKa = 9.52YY53 pKa = 10.77DD54 pKa = 3.55RR55 pKa = 11.84YY56 pKa = 10.86SSNIRR61 pKa = 11.84TSIQYY66 pKa = 9.27PLHH69 pKa = 6.54NSQKK73 pKa = 10.31LNAKK77 pKa = 10.17AFLEE81 pKa = 4.26RR82 pKa = 11.84NGLVPQCQGEE92 pKa = 3.92IDD94 pKa = 4.41NISEE98 pKa = 4.17ANKK101 pKa = 10.48LLLSFKK107 pKa = 10.35KK108 pKa = 10.48LCSHH112 pKa = 6.9NDD114 pKa = 3.43FSSKK118 pKa = 10.41PILPNYY124 pKa = 10.09SNLEE128 pKa = 3.84QWISGQPPSVLKK140 pKa = 10.56ILEE143 pKa = 4.33SEE145 pKa = 4.03EE146 pKa = 4.49GIYY149 pKa = 9.89DD150 pKa = 3.73QRR152 pKa = 11.84FDD154 pKa = 3.54VYY156 pKa = 11.51DD157 pKa = 4.95FIVKK161 pKa = 7.97TIPKK165 pKa = 9.12IDD167 pKa = 3.96LEE169 pKa = 4.76IGAEE173 pKa = 4.03FRR175 pKa = 11.84YY176 pKa = 9.75KK177 pKa = 10.57APQTIAYY184 pKa = 7.76QRR186 pKa = 11.84KK187 pKa = 7.84IFNSVFCPLLKK198 pKa = 10.64EE199 pKa = 3.98FMDD202 pKa = 3.67RR203 pKa = 11.84VEE205 pKa = 4.83IVMDD209 pKa = 4.56KK210 pKa = 11.23NILLYY215 pKa = 11.32NNMSPVEE222 pKa = 3.86FAQYY226 pKa = 7.7FTNIFPVCRR235 pKa = 11.84YY236 pKa = 9.59RR237 pKa = 11.84SLSNFFEE244 pKa = 5.12IDD246 pKa = 3.24FSKK249 pKa = 11.0YY250 pKa = 10.27DD251 pKa = 3.37KK252 pKa = 11.26SQGLVILLFEE262 pKa = 4.32VLVMQEE268 pKa = 4.02FGVPTIYY275 pKa = 10.97LKK277 pKa = 9.97MWVLMHH283 pKa = 7.3RR284 pKa = 11.84FTQIIDD290 pKa = 3.23RR291 pKa = 11.84SNRR294 pKa = 11.84FSAYY298 pKa = 8.93VQYY301 pKa = 10.58QRR303 pKa = 11.84KK304 pKa = 9.85SGDD307 pKa = 3.06AGTWRR312 pKa = 11.84LNTVVQIAILNHH324 pKa = 4.9VFRR327 pKa = 11.84LYY329 pKa = 11.46EE330 pKa = 3.82MVKK333 pKa = 10.42NDD335 pKa = 2.89SCLACFSGDD344 pKa = 4.18DD345 pKa = 3.51SLIFCKK351 pKa = 10.45EE352 pKa = 3.61IVNIDD357 pKa = 3.48QKK359 pKa = 11.76LLFLQTRR366 pKa = 11.84YY367 pKa = 10.43NLEE370 pKa = 4.02AKK372 pKa = 10.19LMNFSVPYY380 pKa = 9.43FCSKK384 pKa = 10.57FLILVDD390 pKa = 3.87NKK392 pKa = 10.28WIFVPDD398 pKa = 3.92TVKK401 pKa = 11.13LIAKK405 pKa = 9.79LGRR408 pKa = 11.84NDD410 pKa = 4.37LVDD413 pKa = 4.58DD414 pKa = 4.15EE415 pKa = 5.71HH416 pKa = 8.23VEE418 pKa = 4.24CYY420 pKa = 10.28RR421 pKa = 11.84ISFEE425 pKa = 5.27DD426 pKa = 3.08NLYY429 pKa = 10.81YY430 pKa = 10.8YY431 pKa = 10.49KK432 pKa = 10.75NCNNWTVISYY442 pKa = 10.08AINDD446 pKa = 3.73RR447 pKa = 11.84YY448 pKa = 9.24KK449 pKa = 10.68TIGEE453 pKa = 3.75HH454 pKa = 6.79DD455 pKa = 3.18IVYY458 pKa = 10.05RR459 pKa = 11.84ALLSSVYY466 pKa = 9.11TKK468 pKa = 10.87SSFRR472 pKa = 11.84RR473 pKa = 11.84LYY475 pKa = 10.57DD476 pKa = 3.22KK477 pKa = 11.29GVGFVKK483 pKa = 10.61GKK485 pKa = 10.47VSSKK489 pKa = 10.52PKK491 pKa = 10.42LDD493 pKa = 3.69FF494 pKa = 4.72
MM1 pKa = 8.01DD2 pKa = 5.02SSNVLQFFFDD12 pKa = 3.72TVLPGSSLEE21 pKa = 4.1FRR23 pKa = 11.84EE24 pKa = 4.03HH25 pKa = 5.67DD26 pKa = 3.37HH27 pKa = 7.47LRR29 pKa = 11.84FEE31 pKa = 5.07YY32 pKa = 7.11EE33 pKa = 3.99TPMKK37 pKa = 10.84AEE39 pKa = 4.63GIMFEE44 pKa = 4.55LSVPTFKK51 pKa = 10.91KK52 pKa = 9.52YY53 pKa = 10.77DD54 pKa = 3.55RR55 pKa = 11.84YY56 pKa = 10.86SSNIRR61 pKa = 11.84TSIQYY66 pKa = 9.27PLHH69 pKa = 6.54NSQKK73 pKa = 10.31LNAKK77 pKa = 10.17AFLEE81 pKa = 4.26RR82 pKa = 11.84NGLVPQCQGEE92 pKa = 3.92IDD94 pKa = 4.41NISEE98 pKa = 4.17ANKK101 pKa = 10.48LLLSFKK107 pKa = 10.35KK108 pKa = 10.48LCSHH112 pKa = 6.9NDD114 pKa = 3.43FSSKK118 pKa = 10.41PILPNYY124 pKa = 10.09SNLEE128 pKa = 3.84QWISGQPPSVLKK140 pKa = 10.56ILEE143 pKa = 4.33SEE145 pKa = 4.03EE146 pKa = 4.49GIYY149 pKa = 9.89DD150 pKa = 3.73QRR152 pKa = 11.84FDD154 pKa = 3.54VYY156 pKa = 11.51DD157 pKa = 4.95FIVKK161 pKa = 7.97TIPKK165 pKa = 9.12IDD167 pKa = 3.96LEE169 pKa = 4.76IGAEE173 pKa = 4.03FRR175 pKa = 11.84YY176 pKa = 9.75KK177 pKa = 10.57APQTIAYY184 pKa = 7.76QRR186 pKa = 11.84KK187 pKa = 7.84IFNSVFCPLLKK198 pKa = 10.64EE199 pKa = 3.98FMDD202 pKa = 3.67RR203 pKa = 11.84VEE205 pKa = 4.83IVMDD209 pKa = 4.56KK210 pKa = 11.23NILLYY215 pKa = 11.32NNMSPVEE222 pKa = 3.86FAQYY226 pKa = 7.7FTNIFPVCRR235 pKa = 11.84YY236 pKa = 9.59RR237 pKa = 11.84SLSNFFEE244 pKa = 5.12IDD246 pKa = 3.24FSKK249 pKa = 11.0YY250 pKa = 10.27DD251 pKa = 3.37KK252 pKa = 11.26SQGLVILLFEE262 pKa = 4.32VLVMQEE268 pKa = 4.02FGVPTIYY275 pKa = 10.97LKK277 pKa = 9.97MWVLMHH283 pKa = 7.3RR284 pKa = 11.84FTQIIDD290 pKa = 3.23RR291 pKa = 11.84SNRR294 pKa = 11.84FSAYY298 pKa = 8.93VQYY301 pKa = 10.58QRR303 pKa = 11.84KK304 pKa = 9.85SGDD307 pKa = 3.06AGTWRR312 pKa = 11.84LNTVVQIAILNHH324 pKa = 4.9VFRR327 pKa = 11.84LYY329 pKa = 11.46EE330 pKa = 3.82MVKK333 pKa = 10.42NDD335 pKa = 2.89SCLACFSGDD344 pKa = 4.18DD345 pKa = 3.51SLIFCKK351 pKa = 10.45EE352 pKa = 3.61IVNIDD357 pKa = 3.48QKK359 pKa = 11.76LLFLQTRR366 pKa = 11.84YY367 pKa = 10.43NLEE370 pKa = 4.02AKK372 pKa = 10.19LMNFSVPYY380 pKa = 9.43FCSKK384 pKa = 10.57FLILVDD390 pKa = 3.87NKK392 pKa = 10.28WIFVPDD398 pKa = 3.92TVKK401 pKa = 11.13LIAKK405 pKa = 9.79LGRR408 pKa = 11.84NDD410 pKa = 4.37LVDD413 pKa = 4.58DD414 pKa = 4.15EE415 pKa = 5.71HH416 pKa = 8.23VEE418 pKa = 4.24CYY420 pKa = 10.28RR421 pKa = 11.84ISFEE425 pKa = 5.27DD426 pKa = 3.08NLYY429 pKa = 10.81YY430 pKa = 10.8YY431 pKa = 10.49KK432 pKa = 10.75NCNNWTVISYY442 pKa = 10.08AINDD446 pKa = 3.73RR447 pKa = 11.84YY448 pKa = 9.24KK449 pKa = 10.68TIGEE453 pKa = 3.75HH454 pKa = 6.79DD455 pKa = 3.18IVYY458 pKa = 10.05RR459 pKa = 11.84ALLSSVYY466 pKa = 9.11TKK468 pKa = 10.87SSFRR472 pKa = 11.84RR473 pKa = 11.84LYY475 pKa = 10.57DD476 pKa = 3.22KK477 pKa = 11.29GVGFVKK483 pKa = 10.61GKK485 pKa = 10.47VSSKK489 pKa = 10.52PKK491 pKa = 10.42LDD493 pKa = 3.69FF494 pKa = 4.72
Molecular weight: 58.01 kDa
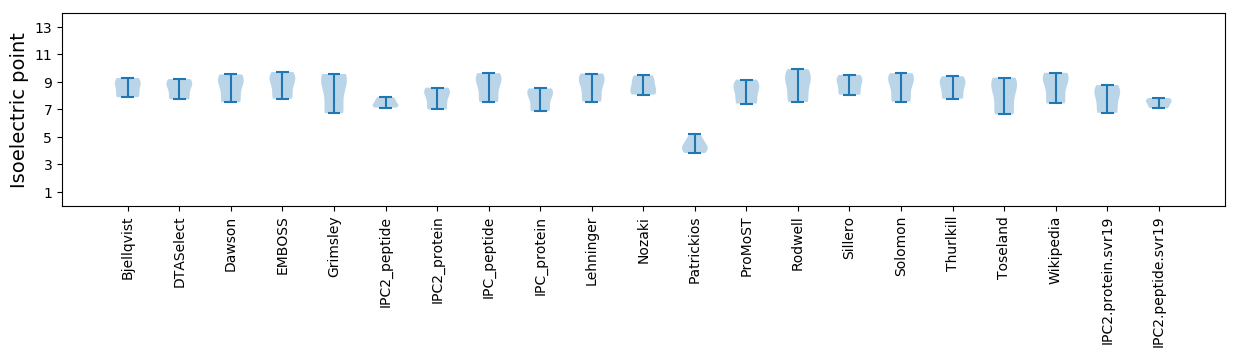
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z2RT34|A0A1Z2RT34_9VIRU Replicase OS=Aedes camptorhynchus negev-like virus OX=2010268 PE=4 SV=1
MM1 pKa = 6.73STKK4 pKa = 10.19PQVTEE9 pKa = 3.79TSEE12 pKa = 4.24VQSGTIIASTQSDD25 pKa = 4.17VKK27 pKa = 10.91SSSPQSTTKK36 pKa = 10.51AVNPFGIKK44 pKa = 9.48ILSGGSVLDD53 pKa = 3.45SSFIQSVVDD62 pKa = 4.23AYY64 pKa = 11.68CNLFSYY70 pKa = 10.6HH71 pKa = 7.22PISLFCMVLSLVYY84 pKa = 10.09FFHH87 pKa = 7.67KK88 pKa = 10.12IIKK91 pKa = 9.49IDD93 pKa = 3.55EE94 pKa = 4.38KK95 pKa = 11.2QDD97 pKa = 3.17IFSTFHH103 pKa = 6.59ANIGKK108 pKa = 8.94AHH110 pKa = 6.51NNSSSIFFKK119 pKa = 11.12GFTTLLAIPASLLVNNKK136 pKa = 7.97LTAATILAFAGPWFAKK152 pKa = 9.78PSSRR156 pKa = 11.84NAIIATILISYY167 pKa = 9.15SLVVTHH173 pKa = 6.54EE174 pKa = 4.31PVSIMILSQAFFLLVEE190 pKa = 4.72LRR192 pKa = 11.84NPLHH196 pKa = 6.68KK197 pKa = 10.9LLVTLFALIVIIIGHH212 pKa = 5.23GQFGAVFKK220 pKa = 11.11
MM1 pKa = 6.73STKK4 pKa = 10.19PQVTEE9 pKa = 3.79TSEE12 pKa = 4.24VQSGTIIASTQSDD25 pKa = 4.17VKK27 pKa = 10.91SSSPQSTTKK36 pKa = 10.51AVNPFGIKK44 pKa = 9.48ILSGGSVLDD53 pKa = 3.45SSFIQSVVDD62 pKa = 4.23AYY64 pKa = 11.68CNLFSYY70 pKa = 10.6HH71 pKa = 7.22PISLFCMVLSLVYY84 pKa = 10.09FFHH87 pKa = 7.67KK88 pKa = 10.12IIKK91 pKa = 9.49IDD93 pKa = 3.55EE94 pKa = 4.38KK95 pKa = 11.2QDD97 pKa = 3.17IFSTFHH103 pKa = 6.59ANIGKK108 pKa = 8.94AHH110 pKa = 6.51NNSSSIFFKK119 pKa = 11.12GFTTLLAIPASLLVNNKK136 pKa = 7.97LTAATILAFAGPWFAKK152 pKa = 9.78PSSRR156 pKa = 11.84NAIIATILISYY167 pKa = 9.15SLVVTHH173 pKa = 6.54EE174 pKa = 4.31PVSIMILSQAFFLLVEE190 pKa = 4.72LRR192 pKa = 11.84NPLHH196 pKa = 6.68KK197 pKa = 10.9LLVTLFALIVIIIGHH212 pKa = 5.23GQFGAVFKK220 pKa = 11.11
Molecular weight: 23.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3492 |
184 |
1899 |
698.4 |
79.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.866 ± 0.465 | 1.919 ± 0.446 |
4.983 ± 0.523 | 5.584 ± 0.691 |
6.93 ± 1.027 | 4.696 ± 0.516 |
1.89 ± 0.21 | 8.104 ± 0.936 |
7.646 ± 0.599 | 10.195 ± 1.219 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.518 ± 0.192 | 6.901 ± 0.63 |
3.666 ± 0.116 | 2.291 ± 0.459 |
4.467 ± 0.69 | 7.446 ± 0.724 |
5.928 ± 0.693 | 7.302 ± 0.341 |
0.601 ± 0.184 | 4.066 ± 0.463 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
