
Streptomyces sp. CB02056
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
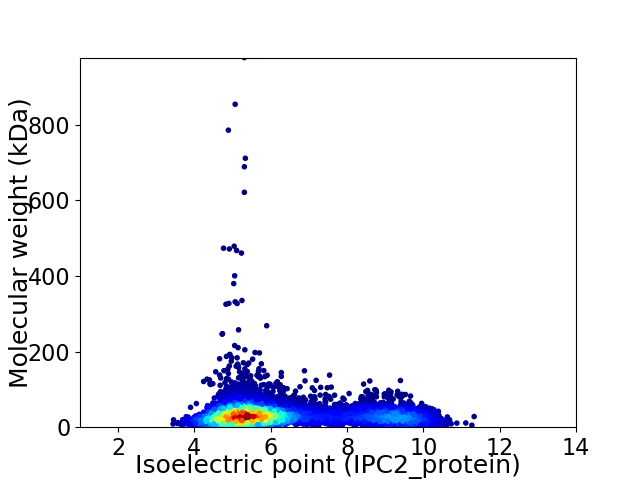
Virtual 2D-PAGE plot for 7266 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
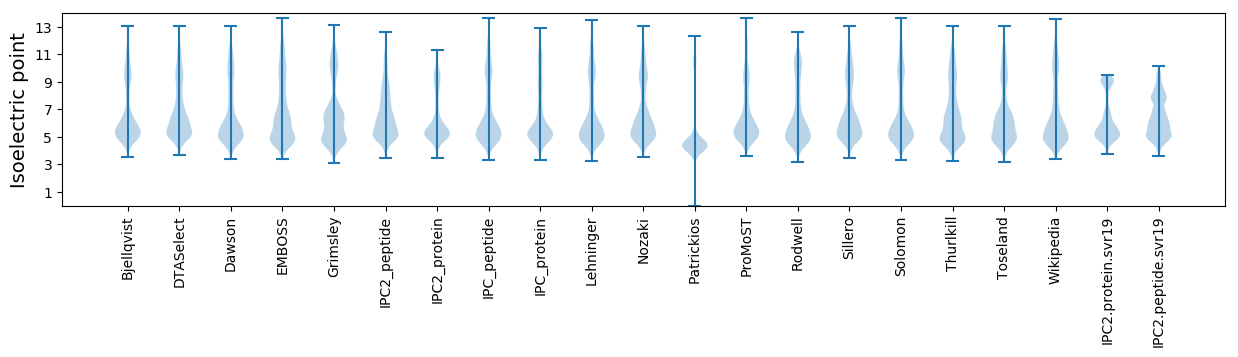
Protein with the lowest isoelectric point:
>tr|A0A1Q4WK48|A0A1Q4WK48_9ACTN Uncharacterized protein OS=Streptomyces sp. CB02056 OX=1703924 GN=AMK13_04920 PE=4 SV=1
MM1 pKa = 7.49PVVTDD6 pKa = 4.33PYY8 pKa = 11.32KK9 pKa = 10.72PGTPCWVDD17 pKa = 4.55LMASDD22 pKa = 3.85QQAALDD28 pKa = 4.19FYY30 pKa = 11.32RR31 pKa = 11.84DD32 pKa = 3.38LFGWQGEE39 pKa = 4.34VGPAEE44 pKa = 3.96FGGYY48 pKa = 8.19AVCMLNGRR56 pKa = 11.84PVAGIMAQAAPEE68 pKa = 4.27GQPTPPVAWTTYY80 pKa = 9.83LASDD84 pKa = 4.1DD85 pKa = 4.33ADD87 pKa = 3.43AARR90 pKa = 11.84ASITEE95 pKa = 3.82AGGTVLYY102 pKa = 10.96DD103 pKa = 3.52PMDD106 pKa = 3.65VGTIGRR112 pKa = 11.84MLVAADD118 pKa = 3.31PTGAVFGVWQAMDD131 pKa = 4.24FIGAGVVNEE140 pKa = 4.37PGALVWNEE148 pKa = 4.27LNTADD153 pKa = 3.62TGAAGRR159 pKa = 11.84FYY161 pKa = 11.0QPALGLRR168 pKa = 11.84PATIQGMDD176 pKa = 3.61GYY178 pKa = 11.1FSLNVADD185 pKa = 4.06RR186 pKa = 11.84TVGGMQAIPGYY197 pKa = 9.62LAAGTPSHH205 pKa = 5.93WMTYY209 pKa = 9.86FSVDD213 pKa = 3.45GADD216 pKa = 3.47STVDD220 pKa = 3.51ALVKK224 pKa = 10.78AGGSVIQPAFDD235 pKa = 3.62MQSGRR240 pKa = 11.84MAVVQDD246 pKa = 3.52PQGAVFAVIEE256 pKa = 4.28APEE259 pKa = 4.0AQMPDD264 pKa = 3.41SPP266 pKa = 4.78
MM1 pKa = 7.49PVVTDD6 pKa = 4.33PYY8 pKa = 11.32KK9 pKa = 10.72PGTPCWVDD17 pKa = 4.55LMASDD22 pKa = 3.85QQAALDD28 pKa = 4.19FYY30 pKa = 11.32RR31 pKa = 11.84DD32 pKa = 3.38LFGWQGEE39 pKa = 4.34VGPAEE44 pKa = 3.96FGGYY48 pKa = 8.19AVCMLNGRR56 pKa = 11.84PVAGIMAQAAPEE68 pKa = 4.27GQPTPPVAWTTYY80 pKa = 9.83LASDD84 pKa = 4.1DD85 pKa = 4.33ADD87 pKa = 3.43AARR90 pKa = 11.84ASITEE95 pKa = 3.82AGGTVLYY102 pKa = 10.96DD103 pKa = 3.52PMDD106 pKa = 3.65VGTIGRR112 pKa = 11.84MLVAADD118 pKa = 3.31PTGAVFGVWQAMDD131 pKa = 4.24FIGAGVVNEE140 pKa = 4.37PGALVWNEE148 pKa = 4.27LNTADD153 pKa = 3.62TGAAGRR159 pKa = 11.84FYY161 pKa = 11.0QPALGLRR168 pKa = 11.84PATIQGMDD176 pKa = 3.61GYY178 pKa = 11.1FSLNVADD185 pKa = 4.06RR186 pKa = 11.84TVGGMQAIPGYY197 pKa = 9.62LAAGTPSHH205 pKa = 5.93WMTYY209 pKa = 9.86FSVDD213 pKa = 3.45GADD216 pKa = 3.47STVDD220 pKa = 3.51ALVKK224 pKa = 10.78AGGSVIQPAFDD235 pKa = 3.62MQSGRR240 pKa = 11.84MAVVQDD246 pKa = 3.52PQGAVFAVIEE256 pKa = 4.28APEE259 pKa = 4.0AQMPDD264 pKa = 3.41SPP266 pKa = 4.78
Molecular weight: 27.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q4W9W1|A0A1Q4W9W1_9ACTN Uncharacterized protein OS=Streptomyces sp. CB02056 OX=1703924 GN=AMK13_15825 PE=4 SV=1
MM1 pKa = 7.49VPGRR5 pKa = 11.84PAAGAVTPPPPLPAVPVRR23 pKa = 11.84PPGRR27 pKa = 11.84GTRR30 pKa = 11.84HH31 pKa = 5.53RR32 pKa = 11.84CGPTVGPPGAHH43 pKa = 6.84PSSARR48 pKa = 11.84RR49 pKa = 11.84PGPARR54 pKa = 11.84PHH56 pKa = 5.68PRR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84PRR63 pKa = 11.84VVPRR67 pKa = 11.84RR68 pKa = 11.84LNPVRR73 pKa = 11.84PLPAGVGPPRR83 pKa = 11.84ARR85 pKa = 11.84LFSAFRR91 pKa = 11.84PRR93 pKa = 11.84RR94 pKa = 11.84LPVVPRR100 pKa = 11.84QPRR103 pKa = 11.84PVRR106 pKa = 11.84PPPAGARR113 pKa = 11.84PSRR116 pKa = 11.84ARR118 pKa = 11.84PSSVRR123 pKa = 11.84RR124 pKa = 11.84PRR126 pKa = 11.84PRR128 pKa = 11.84RR129 pKa = 11.84QHH131 pKa = 6.32RR132 pKa = 11.84FPVVPRR138 pKa = 11.84RR139 pKa = 11.84LSLTRR144 pKa = 11.84PPPVGAGPSRR154 pKa = 11.84APRR157 pKa = 11.84PRR159 pKa = 11.84PRR161 pKa = 11.84RR162 pKa = 11.84QRR164 pKa = 11.84RR165 pKa = 11.84PNSTRR170 pKa = 11.84PLPAGAGPPRR180 pKa = 11.84ARR182 pKa = 11.84PSSVRR187 pKa = 11.84RR188 pKa = 11.84PRR190 pKa = 11.84SRR192 pKa = 11.84RR193 pKa = 11.84QRR195 pKa = 11.84RR196 pKa = 11.84PSPTRR201 pKa = 11.84PLPGRR206 pKa = 11.84AGAWARR212 pKa = 11.84RR213 pKa = 11.84STRR216 pKa = 11.84CRR218 pKa = 11.84RR219 pKa = 11.84RR220 pKa = 11.84PPWRR224 pKa = 11.84GAPLPCSAGADD235 pKa = 3.3RR236 pKa = 11.84SSGSRR241 pKa = 11.84LRR243 pKa = 11.84RR244 pKa = 11.84QARR247 pKa = 11.84PSLPSPRR254 pKa = 11.84RR255 pKa = 3.4
MM1 pKa = 7.49VPGRR5 pKa = 11.84PAAGAVTPPPPLPAVPVRR23 pKa = 11.84PPGRR27 pKa = 11.84GTRR30 pKa = 11.84HH31 pKa = 5.53RR32 pKa = 11.84CGPTVGPPGAHH43 pKa = 6.84PSSARR48 pKa = 11.84RR49 pKa = 11.84PGPARR54 pKa = 11.84PHH56 pKa = 5.68PRR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84PRR63 pKa = 11.84VVPRR67 pKa = 11.84RR68 pKa = 11.84LNPVRR73 pKa = 11.84PLPAGVGPPRR83 pKa = 11.84ARR85 pKa = 11.84LFSAFRR91 pKa = 11.84PRR93 pKa = 11.84RR94 pKa = 11.84LPVVPRR100 pKa = 11.84QPRR103 pKa = 11.84PVRR106 pKa = 11.84PPPAGARR113 pKa = 11.84PSRR116 pKa = 11.84ARR118 pKa = 11.84PSSVRR123 pKa = 11.84RR124 pKa = 11.84PRR126 pKa = 11.84PRR128 pKa = 11.84RR129 pKa = 11.84QHH131 pKa = 6.32RR132 pKa = 11.84FPVVPRR138 pKa = 11.84RR139 pKa = 11.84LSLTRR144 pKa = 11.84PPPVGAGPSRR154 pKa = 11.84APRR157 pKa = 11.84PRR159 pKa = 11.84PRR161 pKa = 11.84RR162 pKa = 11.84QRR164 pKa = 11.84RR165 pKa = 11.84PNSTRR170 pKa = 11.84PLPAGAGPPRR180 pKa = 11.84ARR182 pKa = 11.84PSSVRR187 pKa = 11.84RR188 pKa = 11.84PRR190 pKa = 11.84SRR192 pKa = 11.84RR193 pKa = 11.84QRR195 pKa = 11.84RR196 pKa = 11.84PSPTRR201 pKa = 11.84PLPGRR206 pKa = 11.84AGAWARR212 pKa = 11.84RR213 pKa = 11.84STRR216 pKa = 11.84CRR218 pKa = 11.84RR219 pKa = 11.84RR220 pKa = 11.84PPWRR224 pKa = 11.84GAPLPCSAGADD235 pKa = 3.3RR236 pKa = 11.84SSGSRR241 pKa = 11.84LRR243 pKa = 11.84RR244 pKa = 11.84QARR247 pKa = 11.84PSLPSPRR254 pKa = 11.84RR255 pKa = 3.4
Molecular weight: 28.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2401140 |
36 |
9192 |
330.5 |
35.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.379 ± 0.052 | 0.767 ± 0.008 |
5.834 ± 0.022 | 5.591 ± 0.029 |
2.659 ± 0.016 | 9.712 ± 0.028 |
2.269 ± 0.014 | 2.821 ± 0.022 |
1.798 ± 0.025 | 10.887 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.532 ± 0.013 | 1.698 ± 0.016 |
6.447 ± 0.032 | 2.745 ± 0.017 |
8.116 ± 0.032 | 4.713 ± 0.021 |
6.05 ± 0.025 | 8.455 ± 0.026 |
1.51 ± 0.012 | 2.017 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
