
Dokdonia pacifica
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Dokdonia
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
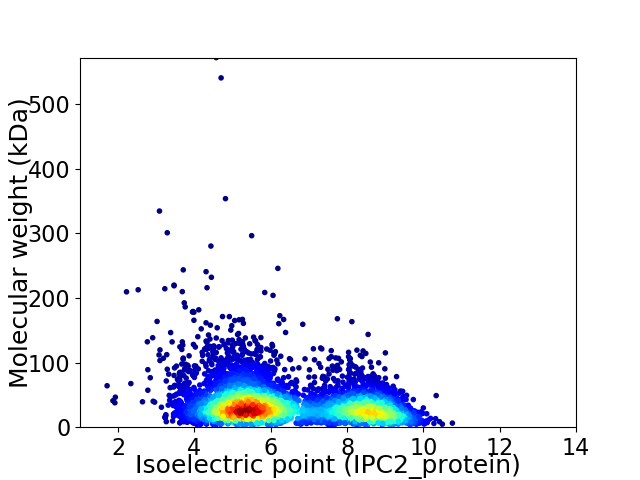
Virtual 2D-PAGE plot for 4883 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A238VLM1|A0A238VLM1_9FLAO FixH protein OS=Dokdonia pacifica OX=1627892 GN=SAMN06265376_10118 PE=4 SV=1
MM1 pKa = 7.06KK2 pKa = 9.7TITYY6 pKa = 10.17RR7 pKa = 11.84RR8 pKa = 11.84ITILLRR14 pKa = 11.84LFTFFLLFPFTIHH27 pKa = 5.68ATIEE31 pKa = 4.02RR32 pKa = 11.84EE33 pKa = 3.89EE34 pKa = 4.15NNISFACTGTVIDD47 pKa = 4.11TFPYY51 pKa = 9.55TEE53 pKa = 4.26SFEE56 pKa = 4.3TGNFDD61 pKa = 3.77FTQNTDD67 pKa = 4.57DD68 pKa = 5.0DD69 pKa = 5.3GDD71 pKa = 3.63WTRR74 pKa = 11.84DD75 pKa = 3.64NNGTPSNNTGPDD87 pKa = 3.49DD88 pKa = 4.97ASDD91 pKa = 3.64GNFYY95 pKa = 11.08LFTEE99 pKa = 5.27ASTSGQGAMGFNASAILTSPCFDD122 pKa = 3.38FTGVGGATFNFDD134 pKa = 2.91YY135 pKa = 11.05HH136 pKa = 6.43MYY138 pKa = 10.71GNAIGQISVEE148 pKa = 4.1VSDD151 pKa = 5.41DD152 pKa = 4.14GGVNWNVLQVISGQQQTSGSAPWITSSINLNAYY185 pKa = 8.92VGNTINVRR193 pKa = 11.84IVGTTGSSFSGDD205 pKa = 3.21MAIDD209 pKa = 3.67NVEE212 pKa = 4.17VTVTPVNPEE221 pKa = 3.52INITGLGNDD230 pKa = 3.55IVSGDD235 pKa = 3.77VTPTVADD242 pKa = 3.7GTDD245 pKa = 3.63FGDD248 pKa = 3.38ITLGLSNSSTFFIEE262 pKa = 3.7NSGILEE268 pKa = 4.68LLLTDD273 pKa = 4.59PNPYY277 pKa = 8.52ITITGTNAGDD287 pKa = 3.43FSIATIPTTPIAPVNGSTPFAIGFTPSAIGDD318 pKa = 3.84RR319 pKa = 11.84FATISIASNDD329 pKa = 3.63SDD331 pKa = 3.71EE332 pKa = 4.37NPYY335 pKa = 10.72VFDD338 pKa = 3.73IMGTGVAPSSEE349 pKa = 4.1INVSGLGNAIMSGDD363 pKa = 4.05VIPSVFDD370 pKa = 3.24GTDD373 pKa = 3.43FGFTTLGTSVSSDD386 pKa = 3.08FVIINSGLLGLDD398 pKa = 3.41LTNASPYY405 pKa = 10.56VNLIGADD412 pKa = 3.24ASQFSISVIPNNTISNNGGTTQFQITYY439 pKa = 8.62TPVSVGVHH447 pKa = 4.93TVTVSIVNNDD457 pKa = 3.37PDD459 pKa = 3.87EE460 pKa = 4.63NPFVFDD466 pKa = 3.18ITGEE470 pKa = 4.05AVVTFVPEE478 pKa = 3.87IEE480 pKa = 4.08ITGNAVEE487 pKa = 4.64IVNGDD492 pKa = 4.24MIPDD496 pKa = 3.37VSDD499 pKa = 3.14ATDD502 pKa = 3.23YY503 pKa = 9.79GTISLGTKK511 pKa = 9.74KK512 pKa = 10.35NVAFTINNVGSGALTLSGPPPYY534 pKa = 9.84LTITGTDD541 pKa = 3.01ASQFSIQTEE550 pKa = 4.09PSSSIAASAATVFQIQYY567 pKa = 10.28NPFGLGTHH575 pKa = 6.35TATVTVVSDD584 pKa = 5.2DD585 pKa = 3.66SDD587 pKa = 3.56EE588 pKa = 4.49PNYY591 pKa = 11.14SFTITGTAIEE601 pKa = 4.55NEE603 pKa = 4.37NPLEE607 pKa = 4.12TPYY610 pKa = 10.86YY611 pKa = 11.11ANFDD615 pKa = 3.78TSDD618 pKa = 4.1DD619 pKa = 3.41NWVASNPGGGSIWTHH634 pKa = 4.39GTNAVEE640 pKa = 4.51VGTEE644 pKa = 3.45GDD646 pKa = 3.11YY647 pKa = 10.48WYY649 pKa = 9.77TDD651 pKa = 2.99TYY653 pKa = 11.75DD654 pKa = 4.97DD655 pKa = 4.12YY656 pKa = 12.04ASNSDD661 pKa = 3.81TYY663 pKa = 9.51VTSPIIDD670 pKa = 3.43LSGYY674 pKa = 10.28QNLTFKK680 pKa = 10.58IDD682 pKa = 2.9IRR684 pKa = 11.84YY685 pKa = 7.86DD686 pKa = 3.33TDD688 pKa = 3.77GDD690 pKa = 3.84TGGDD694 pKa = 3.06DD695 pKa = 4.04GMNIEE700 pKa = 4.34YY701 pKa = 10.68SPDD704 pKa = 3.33GGATWFVLGAYY715 pKa = 9.73ADD717 pKa = 3.85PQVDD721 pKa = 2.69NWYY724 pKa = 10.89NGDD727 pKa = 3.76NVDD730 pKa = 4.48ALGNGIDD737 pKa = 3.5GWAGFSFDD745 pKa = 3.94IVSATKK751 pKa = 10.45SDD753 pKa = 4.9FIQASVDD760 pKa = 3.83LPVEE764 pKa = 4.22LVNNPVARR772 pKa = 11.84FRR774 pKa = 11.84VHH776 pKa = 6.58FASDD780 pKa = 2.98NSQNDD785 pKa = 3.08NGVNFDD791 pKa = 3.64NVFVLGDD798 pKa = 4.6PIVSDD803 pKa = 5.0DD804 pKa = 4.75PIEE807 pKa = 4.68GPAGVYY813 pKa = 9.7TNLKK817 pKa = 9.49LWLKK821 pKa = 6.05TTEE824 pKa = 4.4GVGGASNAGQLAAWEE839 pKa = 4.28DD840 pKa = 3.57QALDD844 pKa = 3.69NNATRR849 pKa = 11.84GDD851 pKa = 3.75SGQQPIYY858 pKa = 10.68YY859 pKa = 10.2DD860 pKa = 5.03DD861 pKa = 3.58INEE864 pKa = 4.54NINYY868 pKa = 9.62NPVVSFNDD876 pKa = 3.83VIDD879 pKa = 4.39SEE881 pKa = 5.03LKK883 pKa = 10.75GKK885 pKa = 10.66GGFNTQDD892 pKa = 2.96YY893 pKa = 8.73WIVMQADD900 pKa = 4.08GSVNSSSPIKK910 pKa = 10.48GVIAGRR916 pKa = 11.84VSRR919 pKa = 11.84DD920 pKa = 3.21QFSEE924 pKa = 4.23DD925 pKa = 4.13GTGLWINPGSLRR937 pKa = 11.84FSGVDD942 pKa = 3.2NIVSHH947 pKa = 5.66MVGATSASASDD958 pKa = 3.96IGDD961 pKa = 3.39GSYY964 pKa = 11.06GRR966 pKa = 11.84AYY968 pKa = 9.74TSSTDD973 pKa = 3.25SYY975 pKa = 11.29DD976 pKa = 3.54NEE978 pKa = 4.64PIILNVKK985 pKa = 9.1VNNAGDD991 pKa = 3.3QTEE994 pKa = 4.29IYY996 pKa = 10.12KK997 pKa = 10.79NGIRR1001 pKa = 11.84IDD1003 pKa = 3.65NYY1005 pKa = 8.97TGKK1008 pKa = 7.46TTGSPSEE1015 pKa = 4.32DD1016 pKa = 2.97LPYY1019 pKa = 10.66INVTNSSYY1027 pKa = 10.92ILGVGRR1033 pKa = 11.84ITVGGTNYY1041 pKa = 10.39DD1042 pKa = 3.34SHH1044 pKa = 8.45FNGKK1048 pKa = 7.78MSEE1051 pKa = 4.35VISYY1055 pKa = 10.41ANPLTIFDD1063 pKa = 3.7QEE1065 pKa = 4.6KK1066 pKa = 8.35IQSYY1070 pKa = 9.8LAIKK1074 pKa = 10.11YY1075 pKa = 9.57GITLHH1080 pKa = 6.53DD1081 pKa = 4.17TNSTSITRR1089 pKa = 11.84LGDD1092 pKa = 3.22QNYY1095 pKa = 9.32IDD1097 pKa = 4.89SDD1099 pKa = 4.03GSTIWNVAAHH1109 pKa = 6.36TGYY1112 pKa = 10.84NYY1114 pKa = 10.53DD1115 pKa = 2.96IAGIGRR1121 pKa = 11.84DD1122 pKa = 3.16ITSGLNQKK1130 pKa = 9.85QSISVNSGAIVTMGLTEE1147 pKa = 4.13VYY1149 pKa = 8.46NTNNEE1154 pKa = 4.16NIANNTNTIVDD1165 pKa = 4.28KK1166 pKa = 11.44NFLMWGNDD1174 pKa = 3.3NKK1176 pKa = 11.0SLNAAASIIVDD1187 pKa = 4.53LSDD1190 pKa = 5.34GIAGLNTVVDD1200 pKa = 3.69ITSVEE1205 pKa = 4.18RR1206 pKa = 11.84SWKK1209 pKa = 9.41VVEE1212 pKa = 4.12TGTVGRR1218 pKa = 11.84VKK1220 pKa = 10.89VSVPEE1225 pKa = 3.94IALSATLDD1233 pKa = 3.66PPGSFFMFISDD1244 pKa = 3.79TPTFNPSSEE1253 pKa = 3.89YY1254 pKa = 10.64RR1255 pKa = 11.84IMLPNGDD1262 pKa = 3.91NVEE1265 pKa = 4.1AEE1267 pKa = 4.16YY1268 pKa = 11.15DD1269 pKa = 3.8FEE1271 pKa = 4.25GTKK1274 pKa = 10.86YY1275 pKa = 9.06ITFGYY1280 pKa = 10.05APDD1283 pKa = 3.76YY1284 pKa = 11.34VFDD1287 pKa = 5.01RR1288 pKa = 11.84SITFNGTTDD1297 pKa = 3.29YY1298 pKa = 11.07MDD1300 pKa = 3.75AQDD1303 pKa = 4.03YY1304 pKa = 11.35LDD1306 pKa = 4.07IEE1308 pKa = 4.79GDD1310 pKa = 3.84FTISTWIKK1318 pKa = 10.41HH1319 pKa = 5.14SSQDD1323 pKa = 3.47YY1324 pKa = 8.8TVLSKK1329 pKa = 11.11NDD1331 pKa = 3.53PAFNEE1336 pKa = 4.91GYY1338 pKa = 9.24EE1339 pKa = 4.16LKK1341 pKa = 10.61INTDD1345 pKa = 3.44RR1346 pKa = 11.84TVSMIWKK1353 pKa = 9.53NGSTQSITSSTQIPNNEE1370 pKa = 3.43WHH1372 pKa = 6.55QIAVSYY1378 pKa = 10.59DD1379 pKa = 3.11GSSASLYY1386 pKa = 9.96IDD1388 pKa = 3.61GVLDD1392 pKa = 3.52VQAPLTAPVTNNEE1405 pKa = 4.14KK1406 pKa = 10.55FLIAAGRR1413 pKa = 11.84GSTPADD1419 pKa = 4.0FYY1421 pKa = 11.25EE1422 pKa = 4.34GTIDD1426 pKa = 4.95EE1427 pKa = 4.35VRR1429 pKa = 11.84VWDD1432 pKa = 4.06AALTANQIRR1441 pKa = 11.84FIMNQEE1447 pKa = 3.53ILEE1450 pKa = 4.25NTNLMVMGEE1459 pKa = 4.91IIPATISKK1467 pKa = 10.48NDD1469 pKa = 3.1IDD1471 pKa = 5.18AIPWDD1476 pKa = 3.88SLDD1479 pKa = 4.14GYY1481 pKa = 11.5FPMTTYY1487 pKa = 11.53AFTNVKK1493 pKa = 9.73GASNNTVVAAIKK1505 pKa = 10.18NLKK1508 pKa = 8.42TVDD1511 pKa = 4.07FQTAPLPYY1519 pKa = 9.89VSQANGNWTNPATWEE1534 pKa = 4.2NGTGFQIPNATSIVDD1549 pKa = 3.27NTVRR1553 pKa = 11.84VDD1555 pKa = 3.13WNIVQTAHH1563 pKa = 6.9NVTTQEE1569 pKa = 3.99NNTVLALDD1577 pKa = 3.75VQTDD1581 pKa = 3.69EE1582 pKa = 5.9LSIEE1586 pKa = 4.07NDD1588 pKa = 3.22SKK1590 pKa = 11.42IEE1592 pKa = 3.8VSHH1595 pKa = 5.33YY1596 pKa = 10.95LKK1598 pKa = 10.85LDD1600 pKa = 3.52GVLDD1604 pKa = 4.01LVGEE1608 pKa = 4.32SQLIQTEE1615 pKa = 4.05NSDD1618 pKa = 4.45LDD1620 pKa = 3.75ATSTGSLEE1628 pKa = 4.22KK1629 pKa = 10.51DD1630 pKa = 3.36QQGTADD1636 pKa = 3.6TYY1638 pKa = 11.63SYY1640 pKa = 10.4NIWAAPVSTINGTQINTNYY1659 pKa = 9.27TVASMMKK1666 pKa = 10.48DD1667 pKa = 2.88GTNPNNPLNMSFTGGLNGAATTPITISSFWMFKK1700 pKa = 9.83YY1701 pKa = 10.91GNGIEE1706 pKa = 4.08QFQPIGSTGNVSVGEE1721 pKa = 4.61GYY1723 pKa = 8.77TMKK1726 pKa = 10.8GPGSGAITDD1735 pKa = 3.65PQNYY1739 pKa = 8.8VFVGKK1744 pKa = 9.59PNNSTDD1750 pKa = 3.38AEE1752 pKa = 4.4EE1753 pKa = 5.14IIVPAIADD1761 pKa = 3.87RR1762 pKa = 11.84DD1763 pKa = 4.12YY1764 pKa = 11.15MVGNPFPSALDD1775 pKa = 3.6ANDD1778 pKa = 5.16FINDD1782 pKa = 3.64NPHH1785 pKa = 7.25LDD1787 pKa = 3.31GTLYY1791 pKa = 10.74FF1792 pKa = 4.81
MM1 pKa = 7.06KK2 pKa = 9.7TITYY6 pKa = 10.17RR7 pKa = 11.84RR8 pKa = 11.84ITILLRR14 pKa = 11.84LFTFFLLFPFTIHH27 pKa = 5.68ATIEE31 pKa = 4.02RR32 pKa = 11.84EE33 pKa = 3.89EE34 pKa = 4.15NNISFACTGTVIDD47 pKa = 4.11TFPYY51 pKa = 9.55TEE53 pKa = 4.26SFEE56 pKa = 4.3TGNFDD61 pKa = 3.77FTQNTDD67 pKa = 4.57DD68 pKa = 5.0DD69 pKa = 5.3GDD71 pKa = 3.63WTRR74 pKa = 11.84DD75 pKa = 3.64NNGTPSNNTGPDD87 pKa = 3.49DD88 pKa = 4.97ASDD91 pKa = 3.64GNFYY95 pKa = 11.08LFTEE99 pKa = 5.27ASTSGQGAMGFNASAILTSPCFDD122 pKa = 3.38FTGVGGATFNFDD134 pKa = 2.91YY135 pKa = 11.05HH136 pKa = 6.43MYY138 pKa = 10.71GNAIGQISVEE148 pKa = 4.1VSDD151 pKa = 5.41DD152 pKa = 4.14GGVNWNVLQVISGQQQTSGSAPWITSSINLNAYY185 pKa = 8.92VGNTINVRR193 pKa = 11.84IVGTTGSSFSGDD205 pKa = 3.21MAIDD209 pKa = 3.67NVEE212 pKa = 4.17VTVTPVNPEE221 pKa = 3.52INITGLGNDD230 pKa = 3.55IVSGDD235 pKa = 3.77VTPTVADD242 pKa = 3.7GTDD245 pKa = 3.63FGDD248 pKa = 3.38ITLGLSNSSTFFIEE262 pKa = 3.7NSGILEE268 pKa = 4.68LLLTDD273 pKa = 4.59PNPYY277 pKa = 8.52ITITGTNAGDD287 pKa = 3.43FSIATIPTTPIAPVNGSTPFAIGFTPSAIGDD318 pKa = 3.84RR319 pKa = 11.84FATISIASNDD329 pKa = 3.63SDD331 pKa = 3.71EE332 pKa = 4.37NPYY335 pKa = 10.72VFDD338 pKa = 3.73IMGTGVAPSSEE349 pKa = 4.1INVSGLGNAIMSGDD363 pKa = 4.05VIPSVFDD370 pKa = 3.24GTDD373 pKa = 3.43FGFTTLGTSVSSDD386 pKa = 3.08FVIINSGLLGLDD398 pKa = 3.41LTNASPYY405 pKa = 10.56VNLIGADD412 pKa = 3.24ASQFSISVIPNNTISNNGGTTQFQITYY439 pKa = 8.62TPVSVGVHH447 pKa = 4.93TVTVSIVNNDD457 pKa = 3.37PDD459 pKa = 3.87EE460 pKa = 4.63NPFVFDD466 pKa = 3.18ITGEE470 pKa = 4.05AVVTFVPEE478 pKa = 3.87IEE480 pKa = 4.08ITGNAVEE487 pKa = 4.64IVNGDD492 pKa = 4.24MIPDD496 pKa = 3.37VSDD499 pKa = 3.14ATDD502 pKa = 3.23YY503 pKa = 9.79GTISLGTKK511 pKa = 9.74KK512 pKa = 10.35NVAFTINNVGSGALTLSGPPPYY534 pKa = 9.84LTITGTDD541 pKa = 3.01ASQFSIQTEE550 pKa = 4.09PSSSIAASAATVFQIQYY567 pKa = 10.28NPFGLGTHH575 pKa = 6.35TATVTVVSDD584 pKa = 5.2DD585 pKa = 3.66SDD587 pKa = 3.56EE588 pKa = 4.49PNYY591 pKa = 11.14SFTITGTAIEE601 pKa = 4.55NEE603 pKa = 4.37NPLEE607 pKa = 4.12TPYY610 pKa = 10.86YY611 pKa = 11.11ANFDD615 pKa = 3.78TSDD618 pKa = 4.1DD619 pKa = 3.41NWVASNPGGGSIWTHH634 pKa = 4.39GTNAVEE640 pKa = 4.51VGTEE644 pKa = 3.45GDD646 pKa = 3.11YY647 pKa = 10.48WYY649 pKa = 9.77TDD651 pKa = 2.99TYY653 pKa = 11.75DD654 pKa = 4.97DD655 pKa = 4.12YY656 pKa = 12.04ASNSDD661 pKa = 3.81TYY663 pKa = 9.51VTSPIIDD670 pKa = 3.43LSGYY674 pKa = 10.28QNLTFKK680 pKa = 10.58IDD682 pKa = 2.9IRR684 pKa = 11.84YY685 pKa = 7.86DD686 pKa = 3.33TDD688 pKa = 3.77GDD690 pKa = 3.84TGGDD694 pKa = 3.06DD695 pKa = 4.04GMNIEE700 pKa = 4.34YY701 pKa = 10.68SPDD704 pKa = 3.33GGATWFVLGAYY715 pKa = 9.73ADD717 pKa = 3.85PQVDD721 pKa = 2.69NWYY724 pKa = 10.89NGDD727 pKa = 3.76NVDD730 pKa = 4.48ALGNGIDD737 pKa = 3.5GWAGFSFDD745 pKa = 3.94IVSATKK751 pKa = 10.45SDD753 pKa = 4.9FIQASVDD760 pKa = 3.83LPVEE764 pKa = 4.22LVNNPVARR772 pKa = 11.84FRR774 pKa = 11.84VHH776 pKa = 6.58FASDD780 pKa = 2.98NSQNDD785 pKa = 3.08NGVNFDD791 pKa = 3.64NVFVLGDD798 pKa = 4.6PIVSDD803 pKa = 5.0DD804 pKa = 4.75PIEE807 pKa = 4.68GPAGVYY813 pKa = 9.7TNLKK817 pKa = 9.49LWLKK821 pKa = 6.05TTEE824 pKa = 4.4GVGGASNAGQLAAWEE839 pKa = 4.28DD840 pKa = 3.57QALDD844 pKa = 3.69NNATRR849 pKa = 11.84GDD851 pKa = 3.75SGQQPIYY858 pKa = 10.68YY859 pKa = 10.2DD860 pKa = 5.03DD861 pKa = 3.58INEE864 pKa = 4.54NINYY868 pKa = 9.62NPVVSFNDD876 pKa = 3.83VIDD879 pKa = 4.39SEE881 pKa = 5.03LKK883 pKa = 10.75GKK885 pKa = 10.66GGFNTQDD892 pKa = 2.96YY893 pKa = 8.73WIVMQADD900 pKa = 4.08GSVNSSSPIKK910 pKa = 10.48GVIAGRR916 pKa = 11.84VSRR919 pKa = 11.84DD920 pKa = 3.21QFSEE924 pKa = 4.23DD925 pKa = 4.13GTGLWINPGSLRR937 pKa = 11.84FSGVDD942 pKa = 3.2NIVSHH947 pKa = 5.66MVGATSASASDD958 pKa = 3.96IGDD961 pKa = 3.39GSYY964 pKa = 11.06GRR966 pKa = 11.84AYY968 pKa = 9.74TSSTDD973 pKa = 3.25SYY975 pKa = 11.29DD976 pKa = 3.54NEE978 pKa = 4.64PIILNVKK985 pKa = 9.1VNNAGDD991 pKa = 3.3QTEE994 pKa = 4.29IYY996 pKa = 10.12KK997 pKa = 10.79NGIRR1001 pKa = 11.84IDD1003 pKa = 3.65NYY1005 pKa = 8.97TGKK1008 pKa = 7.46TTGSPSEE1015 pKa = 4.32DD1016 pKa = 2.97LPYY1019 pKa = 10.66INVTNSSYY1027 pKa = 10.92ILGVGRR1033 pKa = 11.84ITVGGTNYY1041 pKa = 10.39DD1042 pKa = 3.34SHH1044 pKa = 8.45FNGKK1048 pKa = 7.78MSEE1051 pKa = 4.35VISYY1055 pKa = 10.41ANPLTIFDD1063 pKa = 3.7QEE1065 pKa = 4.6KK1066 pKa = 8.35IQSYY1070 pKa = 9.8LAIKK1074 pKa = 10.11YY1075 pKa = 9.57GITLHH1080 pKa = 6.53DD1081 pKa = 4.17TNSTSITRR1089 pKa = 11.84LGDD1092 pKa = 3.22QNYY1095 pKa = 9.32IDD1097 pKa = 4.89SDD1099 pKa = 4.03GSTIWNVAAHH1109 pKa = 6.36TGYY1112 pKa = 10.84NYY1114 pKa = 10.53DD1115 pKa = 2.96IAGIGRR1121 pKa = 11.84DD1122 pKa = 3.16ITSGLNQKK1130 pKa = 9.85QSISVNSGAIVTMGLTEE1147 pKa = 4.13VYY1149 pKa = 8.46NTNNEE1154 pKa = 4.16NIANNTNTIVDD1165 pKa = 4.28KK1166 pKa = 11.44NFLMWGNDD1174 pKa = 3.3NKK1176 pKa = 11.0SLNAAASIIVDD1187 pKa = 4.53LSDD1190 pKa = 5.34GIAGLNTVVDD1200 pKa = 3.69ITSVEE1205 pKa = 4.18RR1206 pKa = 11.84SWKK1209 pKa = 9.41VVEE1212 pKa = 4.12TGTVGRR1218 pKa = 11.84VKK1220 pKa = 10.89VSVPEE1225 pKa = 3.94IALSATLDD1233 pKa = 3.66PPGSFFMFISDD1244 pKa = 3.79TPTFNPSSEE1253 pKa = 3.89YY1254 pKa = 10.64RR1255 pKa = 11.84IMLPNGDD1262 pKa = 3.91NVEE1265 pKa = 4.1AEE1267 pKa = 4.16YY1268 pKa = 11.15DD1269 pKa = 3.8FEE1271 pKa = 4.25GTKK1274 pKa = 10.86YY1275 pKa = 9.06ITFGYY1280 pKa = 10.05APDD1283 pKa = 3.76YY1284 pKa = 11.34VFDD1287 pKa = 5.01RR1288 pKa = 11.84SITFNGTTDD1297 pKa = 3.29YY1298 pKa = 11.07MDD1300 pKa = 3.75AQDD1303 pKa = 4.03YY1304 pKa = 11.35LDD1306 pKa = 4.07IEE1308 pKa = 4.79GDD1310 pKa = 3.84FTISTWIKK1318 pKa = 10.41HH1319 pKa = 5.14SSQDD1323 pKa = 3.47YY1324 pKa = 8.8TVLSKK1329 pKa = 11.11NDD1331 pKa = 3.53PAFNEE1336 pKa = 4.91GYY1338 pKa = 9.24EE1339 pKa = 4.16LKK1341 pKa = 10.61INTDD1345 pKa = 3.44RR1346 pKa = 11.84TVSMIWKK1353 pKa = 9.53NGSTQSITSSTQIPNNEE1370 pKa = 3.43WHH1372 pKa = 6.55QIAVSYY1378 pKa = 10.59DD1379 pKa = 3.11GSSASLYY1386 pKa = 9.96IDD1388 pKa = 3.61GVLDD1392 pKa = 3.52VQAPLTAPVTNNEE1405 pKa = 4.14KK1406 pKa = 10.55FLIAAGRR1413 pKa = 11.84GSTPADD1419 pKa = 4.0FYY1421 pKa = 11.25EE1422 pKa = 4.34GTIDD1426 pKa = 4.95EE1427 pKa = 4.35VRR1429 pKa = 11.84VWDD1432 pKa = 4.06AALTANQIRR1441 pKa = 11.84FIMNQEE1447 pKa = 3.53ILEE1450 pKa = 4.25NTNLMVMGEE1459 pKa = 4.91IIPATISKK1467 pKa = 10.48NDD1469 pKa = 3.1IDD1471 pKa = 5.18AIPWDD1476 pKa = 3.88SLDD1479 pKa = 4.14GYY1481 pKa = 11.5FPMTTYY1487 pKa = 11.53AFTNVKK1493 pKa = 9.73GASNNTVVAAIKK1505 pKa = 10.18NLKK1508 pKa = 8.42TVDD1511 pKa = 4.07FQTAPLPYY1519 pKa = 9.89VSQANGNWTNPATWEE1534 pKa = 4.2NGTGFQIPNATSIVDD1549 pKa = 3.27NTVRR1553 pKa = 11.84VDD1555 pKa = 3.13WNIVQTAHH1563 pKa = 6.9NVTTQEE1569 pKa = 3.99NNTVLALDD1577 pKa = 3.75VQTDD1581 pKa = 3.69EE1582 pKa = 5.9LSIEE1586 pKa = 4.07NDD1588 pKa = 3.22SKK1590 pKa = 11.42IEE1592 pKa = 3.8VSHH1595 pKa = 5.33YY1596 pKa = 10.95LKK1598 pKa = 10.85LDD1600 pKa = 3.52GVLDD1604 pKa = 4.01LVGEE1608 pKa = 4.32SQLIQTEE1615 pKa = 4.05NSDD1618 pKa = 4.45LDD1620 pKa = 3.75ATSTGSLEE1628 pKa = 4.22KK1629 pKa = 10.51DD1630 pKa = 3.36QQGTADD1636 pKa = 3.6TYY1638 pKa = 11.63SYY1640 pKa = 10.4NIWAAPVSTINGTQINTNYY1659 pKa = 9.27TVASMMKK1666 pKa = 10.48DD1667 pKa = 2.88GTNPNNPLNMSFTGGLNGAATTPITISSFWMFKK1700 pKa = 9.83YY1701 pKa = 10.91GNGIEE1706 pKa = 4.08QFQPIGSTGNVSVGEE1721 pKa = 4.61GYY1723 pKa = 8.77TMKK1726 pKa = 10.8GPGSGAITDD1735 pKa = 3.65PQNYY1739 pKa = 8.8VFVGKK1744 pKa = 9.59PNNSTDD1750 pKa = 3.38AEE1752 pKa = 4.4EE1753 pKa = 5.14IIVPAIADD1761 pKa = 3.87RR1762 pKa = 11.84DD1763 pKa = 4.12YY1764 pKa = 11.15MVGNPFPSALDD1775 pKa = 3.6ANDD1778 pKa = 5.16FINDD1782 pKa = 3.64NPHH1785 pKa = 7.25LDD1787 pKa = 3.31GTLYY1791 pKa = 10.74FF1792 pKa = 4.81
Molecular weight: 192.32 kDa
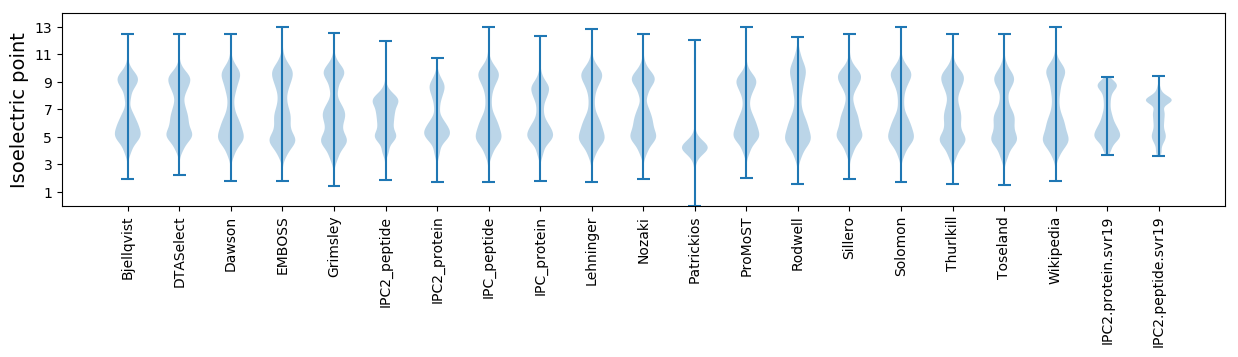
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A239C1Q6|A0A239C1Q6_9FLAO Pimeloyl-ACP methyl ester carboxylesterase OS=Dokdonia pacifica OX=1627892 GN=SAMN06265376_10741 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.19RR22 pKa = 11.84MATANGRR29 pKa = 11.84KK30 pKa = 8.93VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.52LSVSSEE48 pKa = 3.87PRR50 pKa = 11.84HH51 pKa = 5.92KK52 pKa = 10.61KK53 pKa = 9.84
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.19RR22 pKa = 11.84MATANGRR29 pKa = 11.84KK30 pKa = 8.93VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 7.97KK42 pKa = 10.52LSVSSEE48 pKa = 3.87PRR50 pKa = 11.84HH51 pKa = 5.92KK52 pKa = 10.61KK53 pKa = 9.84
Molecular weight: 6.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1653407 |
29 |
5080 |
338.6 |
38.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.367 ± 0.039 | 0.768 ± 0.01 |
5.957 ± 0.043 | 6.492 ± 0.035 |
5.078 ± 0.029 | 6.393 ± 0.051 |
1.854 ± 0.015 | 8.168 ± 0.036 |
6.98 ± 0.052 | 9.085 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.047 ± 0.019 | 5.815 ± 0.036 |
3.489 ± 0.024 | 3.655 ± 0.02 |
3.509 ± 0.027 | 6.508 ± 0.03 |
6.56 ± 0.047 | 6.065 ± 0.03 |
1.047 ± 0.013 | 4.164 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
