
Sanxia sobemo-like virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.02
Get precalculated fractions of proteins
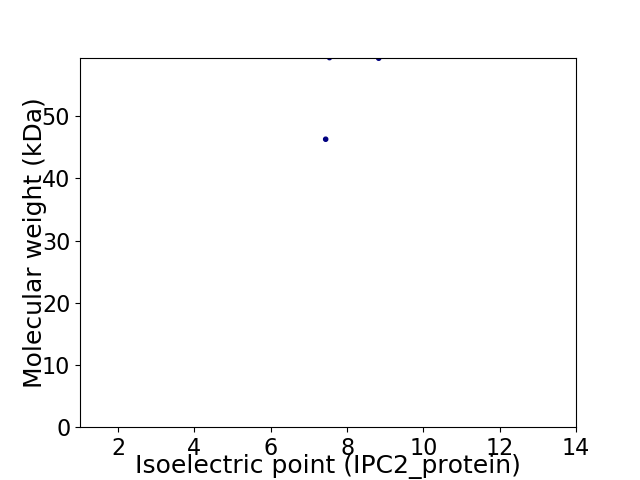
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEU3|A0A1L3KEU3_9VIRU RNA-directed RNA polymerase OS=Sanxia sobemo-like virus 4 OX=1923383 PE=4 SV=1
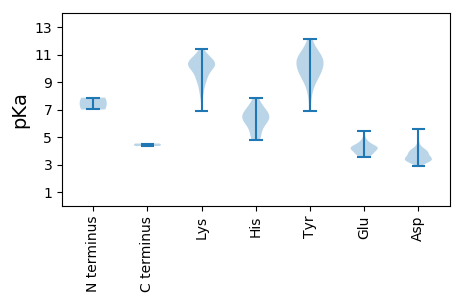
MM1 pKa = 7.86RR2 pKa = 11.84SLAYY6 pKa = 10.23HH7 pKa = 5.35SVLRR11 pKa = 11.84RR12 pKa = 11.84DD13 pKa = 3.39TRR15 pKa = 11.84TDD17 pKa = 3.57GPSGNAKK24 pKa = 8.43TAILNGVLNEE34 pKa = 4.08FMDD37 pKa = 4.65AQWEE41 pKa = 4.34IPTDD45 pKa = 3.96FMTYY49 pKa = 6.87EE50 pKa = 3.86HH51 pKa = 6.75YY52 pKa = 10.89EE53 pKa = 4.07RR54 pKa = 11.84VLSKK58 pKa = 10.23IDD60 pKa = 3.32WTSSPGYY67 pKa = 9.37PYY69 pKa = 9.95MRR71 pKa = 11.84RR72 pKa = 11.84CPTNKK77 pKa = 9.54EE78 pKa = 3.52FFRR81 pKa = 11.84AVDD84 pKa = 3.92GQPSLLRR91 pKa = 11.84KK92 pKa = 7.95NQVWEE97 pKa = 4.02MVQRR101 pKa = 11.84RR102 pKa = 11.84LRR104 pKa = 11.84EE105 pKa = 3.65RR106 pKa = 11.84DD107 pKa = 3.16ADD109 pKa = 4.38PIRR112 pKa = 11.84LFIKK116 pKa = 10.27PEE118 pKa = 3.6PHH120 pKa = 7.13KK121 pKa = 10.59LQKK124 pKa = 10.9LEE126 pKa = 4.18DD127 pKa = 3.32GRR129 pKa = 11.84YY130 pKa = 9.06RR131 pKa = 11.84LISSVSVIDD140 pKa = 4.14QIIDD144 pKa = 3.23HH145 pKa = 6.6MIFGEE150 pKa = 4.23MNEE153 pKa = 4.53KK154 pKa = 10.24LVQNWNYY161 pKa = 10.03VPPKK165 pKa = 10.23IGWSQYY171 pKa = 8.47NGGWRR176 pKa = 11.84SIPTEE181 pKa = 3.7RR182 pKa = 11.84QMALDD187 pKa = 4.12KK188 pKa = 11.19KK189 pKa = 9.49CWDD192 pKa = 3.28WTVQIWVMEE201 pKa = 4.86FILEE205 pKa = 3.93VRR207 pKa = 11.84MRR209 pKa = 11.84LCKK212 pKa = 10.52NLTDD216 pKa = 3.51EE217 pKa = 4.57WKK219 pKa = 10.74DD220 pKa = 3.36LATWRR225 pKa = 11.84YY226 pKa = 9.32KK227 pKa = 10.9SLFVDD232 pKa = 4.06PVFITSGGLMLRR244 pKa = 11.84QRR246 pKa = 11.84NPGVMKK252 pKa = 10.22SGCVLTIGDD261 pKa = 3.8NSIGQVLMHH270 pKa = 6.48HH271 pKa = 6.13RR272 pKa = 11.84VCFEE276 pKa = 3.68NDD278 pKa = 3.16IPPGTIMCMGDD289 pKa = 3.51DD290 pKa = 4.03TLQSVPEE297 pKa = 3.99NTRR300 pKa = 11.84AYY302 pKa = 9.75LDD304 pKa = 3.66TMASFCKK311 pKa = 10.6VKK313 pKa = 10.84DD314 pKa = 3.95PIYY317 pKa = 9.6QSEE320 pKa = 4.19FAGMRR325 pKa = 11.84FYY327 pKa = 10.71PGGRR331 pKa = 11.84VEE333 pKa = 4.35PLYY336 pKa = 10.56KK337 pKa = 10.36GKK339 pKa = 9.49HH340 pKa = 5.42AFTLLHH346 pKa = 6.52VDD348 pKa = 3.92PKK350 pKa = 10.88VLPSLADD357 pKa = 3.51SYY359 pKa = 12.1ALLYY363 pKa = 10.35HH364 pKa = 6.69RR365 pKa = 11.84SSYY368 pKa = 11.22RR369 pKa = 11.84DD370 pKa = 3.29MFEE373 pKa = 5.43DD374 pKa = 4.19LLLKK378 pKa = 10.1MGQKK382 pKa = 10.04VVDD385 pKa = 3.71RR386 pKa = 11.84EE387 pKa = 4.29SRR389 pKa = 11.84DD390 pKa = 3.45VIYY393 pKa = 10.9DD394 pKa = 3.76GFF396 pKa = 4.53
MM1 pKa = 7.86RR2 pKa = 11.84SLAYY6 pKa = 10.23HH7 pKa = 5.35SVLRR11 pKa = 11.84RR12 pKa = 11.84DD13 pKa = 3.39TRR15 pKa = 11.84TDD17 pKa = 3.57GPSGNAKK24 pKa = 8.43TAILNGVLNEE34 pKa = 4.08FMDD37 pKa = 4.65AQWEE41 pKa = 4.34IPTDD45 pKa = 3.96FMTYY49 pKa = 6.87EE50 pKa = 3.86HH51 pKa = 6.75YY52 pKa = 10.89EE53 pKa = 4.07RR54 pKa = 11.84VLSKK58 pKa = 10.23IDD60 pKa = 3.32WTSSPGYY67 pKa = 9.37PYY69 pKa = 9.95MRR71 pKa = 11.84RR72 pKa = 11.84CPTNKK77 pKa = 9.54EE78 pKa = 3.52FFRR81 pKa = 11.84AVDD84 pKa = 3.92GQPSLLRR91 pKa = 11.84KK92 pKa = 7.95NQVWEE97 pKa = 4.02MVQRR101 pKa = 11.84RR102 pKa = 11.84LRR104 pKa = 11.84EE105 pKa = 3.65RR106 pKa = 11.84DD107 pKa = 3.16ADD109 pKa = 4.38PIRR112 pKa = 11.84LFIKK116 pKa = 10.27PEE118 pKa = 3.6PHH120 pKa = 7.13KK121 pKa = 10.59LQKK124 pKa = 10.9LEE126 pKa = 4.18DD127 pKa = 3.32GRR129 pKa = 11.84YY130 pKa = 9.06RR131 pKa = 11.84LISSVSVIDD140 pKa = 4.14QIIDD144 pKa = 3.23HH145 pKa = 6.6MIFGEE150 pKa = 4.23MNEE153 pKa = 4.53KK154 pKa = 10.24LVQNWNYY161 pKa = 10.03VPPKK165 pKa = 10.23IGWSQYY171 pKa = 8.47NGGWRR176 pKa = 11.84SIPTEE181 pKa = 3.7RR182 pKa = 11.84QMALDD187 pKa = 4.12KK188 pKa = 11.19KK189 pKa = 9.49CWDD192 pKa = 3.28WTVQIWVMEE201 pKa = 4.86FILEE205 pKa = 3.93VRR207 pKa = 11.84MRR209 pKa = 11.84LCKK212 pKa = 10.52NLTDD216 pKa = 3.51EE217 pKa = 4.57WKK219 pKa = 10.74DD220 pKa = 3.36LATWRR225 pKa = 11.84YY226 pKa = 9.32KK227 pKa = 10.9SLFVDD232 pKa = 4.06PVFITSGGLMLRR244 pKa = 11.84QRR246 pKa = 11.84NPGVMKK252 pKa = 10.22SGCVLTIGDD261 pKa = 3.8NSIGQVLMHH270 pKa = 6.48HH271 pKa = 6.13RR272 pKa = 11.84VCFEE276 pKa = 3.68NDD278 pKa = 3.16IPPGTIMCMGDD289 pKa = 3.51DD290 pKa = 4.03TLQSVPEE297 pKa = 3.99NTRR300 pKa = 11.84AYY302 pKa = 9.75LDD304 pKa = 3.66TMASFCKK311 pKa = 10.6VKK313 pKa = 10.84DD314 pKa = 3.95PIYY317 pKa = 9.6QSEE320 pKa = 4.19FAGMRR325 pKa = 11.84FYY327 pKa = 10.71PGGRR331 pKa = 11.84VEE333 pKa = 4.35PLYY336 pKa = 10.56KK337 pKa = 10.36GKK339 pKa = 9.49HH340 pKa = 5.42AFTLLHH346 pKa = 6.52VDD348 pKa = 3.92PKK350 pKa = 10.88VLPSLADD357 pKa = 3.51SYY359 pKa = 12.1ALLYY363 pKa = 10.35HH364 pKa = 6.69RR365 pKa = 11.84SSYY368 pKa = 11.22RR369 pKa = 11.84DD370 pKa = 3.29MFEE373 pKa = 5.43DD374 pKa = 4.19LLLKK378 pKa = 10.1MGQKK382 pKa = 10.04VVDD385 pKa = 3.71RR386 pKa = 11.84EE387 pKa = 4.29SRR389 pKa = 11.84DD390 pKa = 3.45VIYY393 pKa = 10.9DD394 pKa = 3.76GFF396 pKa = 4.53
Molecular weight: 46.3 kDa
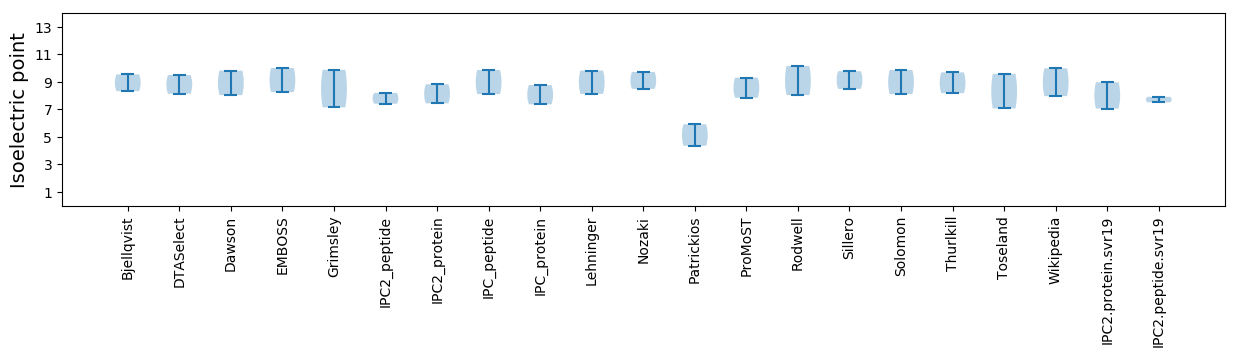
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEU3|A0A1L3KEU3_9VIRU RNA-directed RNA polymerase OS=Sanxia sobemo-like virus 4 OX=1923383 PE=4 SV=1
MM1 pKa = 7.0MWLAITGVTVMTFGFWCFANVLASAGRR28 pKa = 11.84RR29 pKa = 11.84SVQRR33 pKa = 11.84MRR35 pKa = 11.84GIRR38 pKa = 11.84MEE40 pKa = 4.13SVRR43 pKa = 11.84EE44 pKa = 3.8GSRR47 pKa = 11.84FEE49 pKa = 4.0RR50 pKa = 11.84AEE52 pKa = 3.87IPDD55 pKa = 3.41NQVAVRR61 pKa = 11.84IPGTFTDD68 pKa = 3.31EE69 pKa = 4.55HH70 pKa = 7.85IGYY73 pKa = 9.58GVRR76 pKa = 11.84FADD79 pKa = 3.71YY80 pKa = 10.45LVVPRR85 pKa = 11.84HH86 pKa = 4.84VLEE89 pKa = 5.09LSVQGKK95 pKa = 9.1LRR97 pKa = 11.84EE98 pKa = 4.06NVLLVGKK105 pKa = 9.64NGKK108 pKa = 10.11LIVTINATQSRR119 pKa = 11.84GCSDD123 pKa = 3.8LVYY126 pKa = 11.01VNLSAKK132 pKa = 8.13TWSILGAPHH141 pKa = 7.02IKK143 pKa = 8.76WANEE147 pKa = 3.84SKK149 pKa = 10.54NVQVTCTGMPGQSNGRR165 pKa = 11.84IRR167 pKa = 11.84KK168 pKa = 6.91TRR170 pKa = 11.84QEE172 pKa = 3.6WMVSYY177 pKa = 10.42EE178 pKa = 3.9GSTLPGMSGAAYY190 pKa = 9.36MNEE193 pKa = 3.68NLVMGVHH200 pKa = 5.92QGATGAFNMGFSSEE214 pKa = 4.27LVTAEE219 pKa = 3.72MSLLLRR225 pKa = 11.84NEE227 pKa = 4.16DD228 pKa = 3.51TGDD231 pKa = 3.53TNVEE235 pKa = 4.2PKK237 pKa = 10.0QSRR240 pKa = 11.84YY241 pKa = 10.07LSAVQDD247 pKa = 3.4QSFGKK252 pKa = 10.13NFYY255 pKa = 11.0DD256 pKa = 5.55RR257 pKa = 11.84IAALDD262 pKa = 3.9RR263 pKa = 11.84LEE265 pKa = 4.35KK266 pKa = 10.61KK267 pKa = 10.67YY268 pKa = 10.99GAGSDD273 pKa = 3.52WNLDD277 pKa = 2.9GAANYY282 pKa = 8.2DD283 pKa = 3.41QQFDD287 pKa = 3.97FEE289 pKa = 4.67SRR291 pKa = 11.84PRR293 pKa = 11.84PGVSNSSRR301 pKa = 11.84PVRR304 pKa = 11.84VAVPQGGVQLVNHH317 pKa = 6.25NQTGRR322 pKa = 11.84TTLIEE327 pKa = 4.19VQHH330 pKa = 7.33QDD332 pKa = 3.22MLDD335 pKa = 3.17AFANARR341 pKa = 11.84VLDD344 pKa = 3.95RR345 pKa = 11.84LAVLEE350 pKa = 4.16RR351 pKa = 11.84AVGEE355 pKa = 4.41LRR357 pKa = 11.84AQQPPLEE364 pKa = 4.25NDD366 pKa = 3.15RR367 pKa = 11.84LVALEE372 pKa = 3.78QAVRR376 pKa = 11.84DD377 pKa = 3.89LKK379 pKa = 10.39PRR381 pKa = 11.84EE382 pKa = 4.05MPVVMEE388 pKa = 4.67RR389 pKa = 11.84AQPSYY394 pKa = 11.78AEE396 pKa = 4.23VAKK399 pKa = 9.18TAPSEE404 pKa = 3.93KK405 pKa = 10.17KK406 pKa = 9.35PRR408 pKa = 11.84KK409 pKa = 9.64KK410 pKa = 10.38YY411 pKa = 10.41ACKK414 pKa = 10.16LCNIVTRR421 pKa = 11.84TQMRR425 pKa = 11.84LDD427 pKa = 3.2NHH429 pKa = 6.3IASNHH434 pKa = 4.81KK435 pKa = 8.56TGQVQMEE442 pKa = 4.3SAIPSDD448 pKa = 3.37TGKK451 pKa = 10.16SGKK454 pKa = 8.97IIKK457 pKa = 9.32MGSFLEE463 pKa = 4.4KK464 pKa = 10.15RR465 pKa = 11.84SSSPRR470 pKa = 11.84NKK472 pKa = 10.06SNQSSRR478 pKa = 11.84SSRR481 pKa = 11.84FSEE484 pKa = 4.38KK485 pKa = 10.18KK486 pKa = 9.15NHH488 pKa = 6.11SPLPEE493 pKa = 4.73GSLSEE498 pKa = 4.41MIASQRR504 pKa = 11.84SISADD509 pKa = 3.2LKK511 pKa = 10.37EE512 pKa = 4.26LLKK515 pKa = 11.43VMVGQSSAMTQKK527 pKa = 10.62CGVSPTTQCC536 pKa = 4.35
MM1 pKa = 7.0MWLAITGVTVMTFGFWCFANVLASAGRR28 pKa = 11.84RR29 pKa = 11.84SVQRR33 pKa = 11.84MRR35 pKa = 11.84GIRR38 pKa = 11.84MEE40 pKa = 4.13SVRR43 pKa = 11.84EE44 pKa = 3.8GSRR47 pKa = 11.84FEE49 pKa = 4.0RR50 pKa = 11.84AEE52 pKa = 3.87IPDD55 pKa = 3.41NQVAVRR61 pKa = 11.84IPGTFTDD68 pKa = 3.31EE69 pKa = 4.55HH70 pKa = 7.85IGYY73 pKa = 9.58GVRR76 pKa = 11.84FADD79 pKa = 3.71YY80 pKa = 10.45LVVPRR85 pKa = 11.84HH86 pKa = 4.84VLEE89 pKa = 5.09LSVQGKK95 pKa = 9.1LRR97 pKa = 11.84EE98 pKa = 4.06NVLLVGKK105 pKa = 9.64NGKK108 pKa = 10.11LIVTINATQSRR119 pKa = 11.84GCSDD123 pKa = 3.8LVYY126 pKa = 11.01VNLSAKK132 pKa = 8.13TWSILGAPHH141 pKa = 7.02IKK143 pKa = 8.76WANEE147 pKa = 3.84SKK149 pKa = 10.54NVQVTCTGMPGQSNGRR165 pKa = 11.84IRR167 pKa = 11.84KK168 pKa = 6.91TRR170 pKa = 11.84QEE172 pKa = 3.6WMVSYY177 pKa = 10.42EE178 pKa = 3.9GSTLPGMSGAAYY190 pKa = 9.36MNEE193 pKa = 3.68NLVMGVHH200 pKa = 5.92QGATGAFNMGFSSEE214 pKa = 4.27LVTAEE219 pKa = 3.72MSLLLRR225 pKa = 11.84NEE227 pKa = 4.16DD228 pKa = 3.51TGDD231 pKa = 3.53TNVEE235 pKa = 4.2PKK237 pKa = 10.0QSRR240 pKa = 11.84YY241 pKa = 10.07LSAVQDD247 pKa = 3.4QSFGKK252 pKa = 10.13NFYY255 pKa = 11.0DD256 pKa = 5.55RR257 pKa = 11.84IAALDD262 pKa = 3.9RR263 pKa = 11.84LEE265 pKa = 4.35KK266 pKa = 10.61KK267 pKa = 10.67YY268 pKa = 10.99GAGSDD273 pKa = 3.52WNLDD277 pKa = 2.9GAANYY282 pKa = 8.2DD283 pKa = 3.41QQFDD287 pKa = 3.97FEE289 pKa = 4.67SRR291 pKa = 11.84PRR293 pKa = 11.84PGVSNSSRR301 pKa = 11.84PVRR304 pKa = 11.84VAVPQGGVQLVNHH317 pKa = 6.25NQTGRR322 pKa = 11.84TTLIEE327 pKa = 4.19VQHH330 pKa = 7.33QDD332 pKa = 3.22MLDD335 pKa = 3.17AFANARR341 pKa = 11.84VLDD344 pKa = 3.95RR345 pKa = 11.84LAVLEE350 pKa = 4.16RR351 pKa = 11.84AVGEE355 pKa = 4.41LRR357 pKa = 11.84AQQPPLEE364 pKa = 4.25NDD366 pKa = 3.15RR367 pKa = 11.84LVALEE372 pKa = 3.78QAVRR376 pKa = 11.84DD377 pKa = 3.89LKK379 pKa = 10.39PRR381 pKa = 11.84EE382 pKa = 4.05MPVVMEE388 pKa = 4.67RR389 pKa = 11.84AQPSYY394 pKa = 11.78AEE396 pKa = 4.23VAKK399 pKa = 9.18TAPSEE404 pKa = 3.93KK405 pKa = 10.17KK406 pKa = 9.35PRR408 pKa = 11.84KK409 pKa = 9.64KK410 pKa = 10.38YY411 pKa = 10.41ACKK414 pKa = 10.16LCNIVTRR421 pKa = 11.84TQMRR425 pKa = 11.84LDD427 pKa = 3.2NHH429 pKa = 6.3IASNHH434 pKa = 4.81KK435 pKa = 8.56TGQVQMEE442 pKa = 4.3SAIPSDD448 pKa = 3.37TGKK451 pKa = 10.16SGKK454 pKa = 8.97IIKK457 pKa = 9.32MGSFLEE463 pKa = 4.4KK464 pKa = 10.15RR465 pKa = 11.84SSSPRR470 pKa = 11.84NKK472 pKa = 10.06SNQSSRR478 pKa = 11.84SSRR481 pKa = 11.84FSEE484 pKa = 4.38KK485 pKa = 10.18KK486 pKa = 9.15NHH488 pKa = 6.11SPLPEE493 pKa = 4.73GSLSEE498 pKa = 4.41MIASQRR504 pKa = 11.84SISADD509 pKa = 3.2LKK511 pKa = 10.37EE512 pKa = 4.26LLKK515 pKa = 11.43VMVGQSSAMTQKK527 pKa = 10.62CGVSPTTQCC536 pKa = 4.35
Molecular weight: 59.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
932 |
396 |
536 |
466.0 |
52.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.901 ± 1.56 | 1.502 ± 0.175 |
5.365 ± 1.292 | 5.687 ± 0.253 |
3.326 ± 0.471 | 6.867 ± 0.532 |
1.931 ± 0.225 | 4.292 ± 0.667 |
5.472 ± 0.055 | 8.155 ± 0.451 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.292 ± 0.334 | 4.506 ± 0.64 |
4.828 ± 0.48 | 4.828 ± 0.686 |
7.725 ± 0.068 | 7.94 ± 1.239 |
4.828 ± 0.187 | 7.725 ± 0.598 |
1.824 ± 0.629 | 3.004 ± 0.85 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
