
Wuhan horsefly Virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Horwuvirus; Horsefly horwuvirus
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
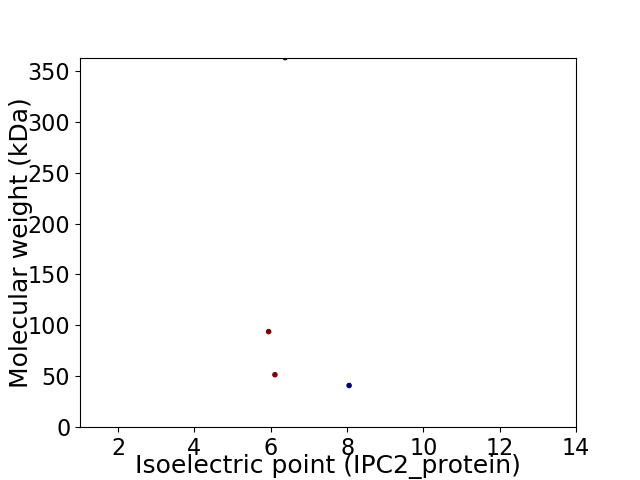
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KY39|A0A0B5KY39_9VIRU Glycoprotein OS=Wuhan horsefly Virus OX=1608139 GN=G PE=4 SV=1
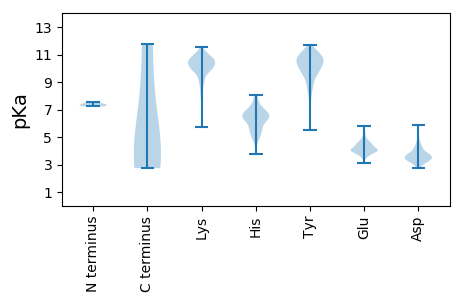
MM1 pKa = 7.32GCLRR5 pKa = 11.84FIIIVISILIVNKK18 pKa = 9.82RR19 pKa = 11.84VCSMEE24 pKa = 3.71DD25 pKa = 3.01RR26 pKa = 11.84EE27 pKa = 4.91IIVHH31 pKa = 5.8LTNKK35 pKa = 9.37VVEE38 pKa = 4.62DD39 pKa = 3.54ATIQDD44 pKa = 3.35KK45 pKa = 10.44TEE47 pKa = 3.65FLIYY51 pKa = 10.76LNDD54 pKa = 3.6ITSLDD59 pKa = 3.47PWYY62 pKa = 10.27HH63 pKa = 6.41QIGCKK68 pKa = 9.04TNNCGLRR75 pKa = 11.84YY76 pKa = 10.21NNNSKK81 pKa = 10.55QYY83 pKa = 10.93DD84 pKa = 3.28IALSQKK90 pKa = 9.9NVNLGWFYY98 pKa = 11.33GSKK101 pKa = 10.38ALKK104 pKa = 10.26DD105 pKa = 3.39KK106 pKa = 9.13PTYY109 pKa = 10.2PGVDD113 pKa = 3.45CLTPFIVINDD123 pKa = 4.21KK124 pKa = 9.32ITYY127 pKa = 9.63VPEE130 pKa = 4.15LLVKK134 pKa = 9.48FIVSICSSDD143 pKa = 3.33RR144 pKa = 11.84GRR146 pKa = 11.84KK147 pKa = 8.9CKK149 pKa = 10.64ACDD152 pKa = 3.6NNCVYY157 pKa = 10.65QSDD160 pKa = 5.19LINFGLMRR168 pKa = 11.84TNCQLVIVEE177 pKa = 4.59NNTTIPYY184 pKa = 10.31DD185 pKa = 4.09VIDD188 pKa = 3.62SPYY191 pKa = 9.6TYY193 pKa = 10.54EE194 pKa = 4.3DD195 pKa = 3.17KK196 pKa = 11.07CVNGEE201 pKa = 3.5ICMDD205 pKa = 3.5STIEE209 pKa = 4.07SLCYY213 pKa = 10.12SGSCTEE219 pKa = 4.58IKK221 pKa = 10.6DD222 pKa = 3.91NQPEE226 pKa = 4.19KK227 pKa = 10.53CVRR230 pKa = 11.84IPWKK234 pKa = 9.26YY235 pKa = 8.35PKK237 pKa = 10.54YY238 pKa = 10.68GIIEE242 pKa = 4.1VKK244 pKa = 10.28LQNNKK249 pKa = 8.07TSQMTCKK256 pKa = 9.78IDD258 pKa = 4.24DD259 pKa = 3.87FCTLIRR265 pKa = 11.84SYY267 pKa = 11.04TYY269 pKa = 7.33FTPLLHH275 pKa = 7.01PYY277 pKa = 10.23CSSTKK282 pKa = 10.06EE283 pKa = 3.63KK284 pKa = 11.04VIFYY288 pKa = 9.11TLFIMLIWSVLILTIQFKK306 pKa = 10.34LYY308 pKa = 10.14KK309 pKa = 10.02VLYY312 pKa = 8.61PCYY315 pKa = 10.51KK316 pKa = 9.68LTSCLKK322 pKa = 9.96FEE324 pKa = 4.08IMKK327 pKa = 10.25YY328 pKa = 9.77LYY330 pKa = 8.88EE331 pKa = 3.9RR332 pKa = 11.84HH333 pKa = 6.23SIEE336 pKa = 4.88IEE338 pKa = 3.8MDD340 pKa = 3.52VLNSDD345 pKa = 3.4GSARR349 pKa = 11.84KK350 pKa = 9.22GKK352 pKa = 10.11KK353 pKa = 9.09SKK355 pKa = 10.27SWLKK359 pKa = 10.47QLLLLIFCLQRR370 pKa = 11.84VEE372 pKa = 4.56SSCDD376 pKa = 3.18FAFPIHH382 pKa = 7.09SSTSVCINGSCDD394 pKa = 2.98EE395 pKa = 4.45MEE397 pKa = 4.32FMSYY401 pKa = 10.09HH402 pKa = 5.53VPSGNLHH409 pKa = 5.5ICGNNIDD416 pKa = 3.84IYY418 pKa = 11.02IMKK421 pKa = 9.5TYY423 pKa = 10.08LRR425 pKa = 11.84CVPDD429 pKa = 3.19ILYY432 pKa = 7.37YY433 pKa = 10.32TPITIIKK440 pKa = 10.24ASRR443 pKa = 11.84TWNCLWAGDD452 pKa = 4.27CTNTCNSSVINHH464 pKa = 7.2EE465 pKa = 4.22INEE468 pKa = 3.98KK469 pKa = 10.25DD470 pKa = 3.36QYY472 pKa = 10.45VHH474 pKa = 6.7LRR476 pKa = 11.84SCEE479 pKa = 3.96KK480 pKa = 10.69VPGSGNVCRR489 pKa = 11.84YY490 pKa = 10.7DD491 pKa = 5.27DD492 pKa = 3.5ICVWRR497 pKa = 11.84NIFIAGKK504 pKa = 10.28DD505 pKa = 3.32NGLKK509 pKa = 10.04FLRR512 pKa = 11.84VSSCTEE518 pKa = 3.37HH519 pKa = 8.03LYY521 pKa = 10.88FSSLLIKK528 pKa = 10.51SEE530 pKa = 3.99NQSFNLMAEE539 pKa = 3.99HH540 pKa = 6.14GTLYY544 pKa = 11.15NLGDD548 pKa = 3.7KK549 pKa = 11.03LSFQIINTNSFRR561 pKa = 11.84KK562 pKa = 9.64DD563 pKa = 3.29LKK565 pKa = 11.02LLEE568 pKa = 5.27LGDD571 pKa = 3.72KK572 pKa = 11.0GYY574 pKa = 11.0LGDD577 pKa = 4.6ANNLHH582 pKa = 6.58EE583 pKa = 4.75SVRR586 pKa = 11.84HH587 pKa = 5.14KK588 pKa = 10.44PGSIQCEE595 pKa = 3.97DD596 pKa = 3.05SDD598 pKa = 4.56IYY600 pKa = 11.45SNCRR604 pKa = 11.84WNKK607 pKa = 7.8EE608 pKa = 3.79SEE610 pKa = 4.14MFDD613 pKa = 3.35GGFQEE618 pKa = 5.64HH619 pKa = 6.21YY620 pKa = 10.87LVDD623 pKa = 4.5NEE625 pKa = 5.3DD626 pKa = 3.16KK627 pKa = 10.73PQRR630 pKa = 11.84LDD632 pKa = 3.37EE633 pKa = 4.44AFKK636 pKa = 10.36KK637 pKa = 10.25ARR639 pKa = 11.84HH640 pKa = 5.82IVTADD645 pKa = 3.05GVEE648 pKa = 4.13IEE650 pKa = 4.61NLPFTLMVSNRR661 pKa = 11.84NHH663 pKa = 6.65LKK665 pKa = 10.86DD666 pKa = 5.14KK667 pKa = 10.81ISLDD671 pKa = 3.65LGEE674 pKa = 4.32LTIANGFVEE683 pKa = 4.45GLSYY687 pKa = 11.28LDD689 pKa = 4.52NKK691 pKa = 10.64CKK693 pKa = 10.17MSFTYY698 pKa = 10.7SLTEE702 pKa = 3.51QMVIQISSPIITSSILKK719 pKa = 9.77LDD721 pKa = 3.48KK722 pKa = 11.1AKK724 pKa = 10.7EE725 pKa = 4.17SEE727 pKa = 4.04LSINCTCLYY736 pKa = 10.77SEE738 pKa = 5.01VDD740 pKa = 3.25TTQITFSAYY749 pKa = 9.76HH750 pKa = 6.4LSKK753 pKa = 9.96TYY755 pKa = 8.8TFKK758 pKa = 10.98TNLSHH763 pKa = 7.87PIGHH767 pKa = 6.15FTQYY771 pKa = 11.29HH772 pKa = 6.99DD773 pKa = 3.5IFLGKK778 pKa = 10.15SLGGYY783 pKa = 10.24DD784 pKa = 3.66YY785 pKa = 11.25FSGLSHH791 pKa = 7.14FFSSHH796 pKa = 5.56SDD798 pKa = 3.84FIYY801 pKa = 10.62SIVGVIIVLFLLKK814 pKa = 10.74LFLCC818 pKa = 5.11
MM1 pKa = 7.32GCLRR5 pKa = 11.84FIIIVISILIVNKK18 pKa = 9.82RR19 pKa = 11.84VCSMEE24 pKa = 3.71DD25 pKa = 3.01RR26 pKa = 11.84EE27 pKa = 4.91IIVHH31 pKa = 5.8LTNKK35 pKa = 9.37VVEE38 pKa = 4.62DD39 pKa = 3.54ATIQDD44 pKa = 3.35KK45 pKa = 10.44TEE47 pKa = 3.65FLIYY51 pKa = 10.76LNDD54 pKa = 3.6ITSLDD59 pKa = 3.47PWYY62 pKa = 10.27HH63 pKa = 6.41QIGCKK68 pKa = 9.04TNNCGLRR75 pKa = 11.84YY76 pKa = 10.21NNNSKK81 pKa = 10.55QYY83 pKa = 10.93DD84 pKa = 3.28IALSQKK90 pKa = 9.9NVNLGWFYY98 pKa = 11.33GSKK101 pKa = 10.38ALKK104 pKa = 10.26DD105 pKa = 3.39KK106 pKa = 9.13PTYY109 pKa = 10.2PGVDD113 pKa = 3.45CLTPFIVINDD123 pKa = 4.21KK124 pKa = 9.32ITYY127 pKa = 9.63VPEE130 pKa = 4.15LLVKK134 pKa = 9.48FIVSICSSDD143 pKa = 3.33RR144 pKa = 11.84GRR146 pKa = 11.84KK147 pKa = 8.9CKK149 pKa = 10.64ACDD152 pKa = 3.6NNCVYY157 pKa = 10.65QSDD160 pKa = 5.19LINFGLMRR168 pKa = 11.84TNCQLVIVEE177 pKa = 4.59NNTTIPYY184 pKa = 10.31DD185 pKa = 4.09VIDD188 pKa = 3.62SPYY191 pKa = 9.6TYY193 pKa = 10.54EE194 pKa = 4.3DD195 pKa = 3.17KK196 pKa = 11.07CVNGEE201 pKa = 3.5ICMDD205 pKa = 3.5STIEE209 pKa = 4.07SLCYY213 pKa = 10.12SGSCTEE219 pKa = 4.58IKK221 pKa = 10.6DD222 pKa = 3.91NQPEE226 pKa = 4.19KK227 pKa = 10.53CVRR230 pKa = 11.84IPWKK234 pKa = 9.26YY235 pKa = 8.35PKK237 pKa = 10.54YY238 pKa = 10.68GIIEE242 pKa = 4.1VKK244 pKa = 10.28LQNNKK249 pKa = 8.07TSQMTCKK256 pKa = 9.78IDD258 pKa = 4.24DD259 pKa = 3.87FCTLIRR265 pKa = 11.84SYY267 pKa = 11.04TYY269 pKa = 7.33FTPLLHH275 pKa = 7.01PYY277 pKa = 10.23CSSTKK282 pKa = 10.06EE283 pKa = 3.63KK284 pKa = 11.04VIFYY288 pKa = 9.11TLFIMLIWSVLILTIQFKK306 pKa = 10.34LYY308 pKa = 10.14KK309 pKa = 10.02VLYY312 pKa = 8.61PCYY315 pKa = 10.51KK316 pKa = 9.68LTSCLKK322 pKa = 9.96FEE324 pKa = 4.08IMKK327 pKa = 10.25YY328 pKa = 9.77LYY330 pKa = 8.88EE331 pKa = 3.9RR332 pKa = 11.84HH333 pKa = 6.23SIEE336 pKa = 4.88IEE338 pKa = 3.8MDD340 pKa = 3.52VLNSDD345 pKa = 3.4GSARR349 pKa = 11.84KK350 pKa = 9.22GKK352 pKa = 10.11KK353 pKa = 9.09SKK355 pKa = 10.27SWLKK359 pKa = 10.47QLLLLIFCLQRR370 pKa = 11.84VEE372 pKa = 4.56SSCDD376 pKa = 3.18FAFPIHH382 pKa = 7.09SSTSVCINGSCDD394 pKa = 2.98EE395 pKa = 4.45MEE397 pKa = 4.32FMSYY401 pKa = 10.09HH402 pKa = 5.53VPSGNLHH409 pKa = 5.5ICGNNIDD416 pKa = 3.84IYY418 pKa = 11.02IMKK421 pKa = 9.5TYY423 pKa = 10.08LRR425 pKa = 11.84CVPDD429 pKa = 3.19ILYY432 pKa = 7.37YY433 pKa = 10.32TPITIIKK440 pKa = 10.24ASRR443 pKa = 11.84TWNCLWAGDD452 pKa = 4.27CTNTCNSSVINHH464 pKa = 7.2EE465 pKa = 4.22INEE468 pKa = 3.98KK469 pKa = 10.25DD470 pKa = 3.36QYY472 pKa = 10.45VHH474 pKa = 6.7LRR476 pKa = 11.84SCEE479 pKa = 3.96KK480 pKa = 10.69VPGSGNVCRR489 pKa = 11.84YY490 pKa = 10.7DD491 pKa = 5.27DD492 pKa = 3.5ICVWRR497 pKa = 11.84NIFIAGKK504 pKa = 10.28DD505 pKa = 3.32NGLKK509 pKa = 10.04FLRR512 pKa = 11.84VSSCTEE518 pKa = 3.37HH519 pKa = 8.03LYY521 pKa = 10.88FSSLLIKK528 pKa = 10.51SEE530 pKa = 3.99NQSFNLMAEE539 pKa = 3.99HH540 pKa = 6.14GTLYY544 pKa = 11.15NLGDD548 pKa = 3.7KK549 pKa = 11.03LSFQIINTNSFRR561 pKa = 11.84KK562 pKa = 9.64DD563 pKa = 3.29LKK565 pKa = 11.02LLEE568 pKa = 5.27LGDD571 pKa = 3.72KK572 pKa = 11.0GYY574 pKa = 11.0LGDD577 pKa = 4.6ANNLHH582 pKa = 6.58EE583 pKa = 4.75SVRR586 pKa = 11.84HH587 pKa = 5.14KK588 pKa = 10.44PGSIQCEE595 pKa = 3.97DD596 pKa = 3.05SDD598 pKa = 4.56IYY600 pKa = 11.45SNCRR604 pKa = 11.84WNKK607 pKa = 7.8EE608 pKa = 3.79SEE610 pKa = 4.14MFDD613 pKa = 3.35GGFQEE618 pKa = 5.64HH619 pKa = 6.21YY620 pKa = 10.87LVDD623 pKa = 4.5NEE625 pKa = 5.3DD626 pKa = 3.16KK627 pKa = 10.73PQRR630 pKa = 11.84LDD632 pKa = 3.37EE633 pKa = 4.44AFKK636 pKa = 10.36KK637 pKa = 10.25ARR639 pKa = 11.84HH640 pKa = 5.82IVTADD645 pKa = 3.05GVEE648 pKa = 4.13IEE650 pKa = 4.61NLPFTLMVSNRR661 pKa = 11.84NHH663 pKa = 6.65LKK665 pKa = 10.86DD666 pKa = 5.14KK667 pKa = 10.81ISLDD671 pKa = 3.65LGEE674 pKa = 4.32LTIANGFVEE683 pKa = 4.45GLSYY687 pKa = 11.28LDD689 pKa = 4.52NKK691 pKa = 10.64CKK693 pKa = 10.17MSFTYY698 pKa = 10.7SLTEE702 pKa = 3.51QMVIQISSPIITSSILKK719 pKa = 9.77LDD721 pKa = 3.48KK722 pKa = 11.1AKK724 pKa = 10.7EE725 pKa = 4.17SEE727 pKa = 4.04LSINCTCLYY736 pKa = 10.77SEE738 pKa = 5.01VDD740 pKa = 3.25TTQITFSAYY749 pKa = 9.76HH750 pKa = 6.4LSKK753 pKa = 9.96TYY755 pKa = 8.8TFKK758 pKa = 10.98TNLSHH763 pKa = 7.87PIGHH767 pKa = 6.15FTQYY771 pKa = 11.29HH772 pKa = 6.99DD773 pKa = 3.5IFLGKK778 pKa = 10.15SLGGYY783 pKa = 10.24DD784 pKa = 3.66YY785 pKa = 11.25FSGLSHH791 pKa = 7.14FFSSHH796 pKa = 5.56SDD798 pKa = 3.84FIYY801 pKa = 10.62SIVGVIIVLFLLKK814 pKa = 10.74LFLCC818 pKa = 5.11
Molecular weight: 93.85 kDa
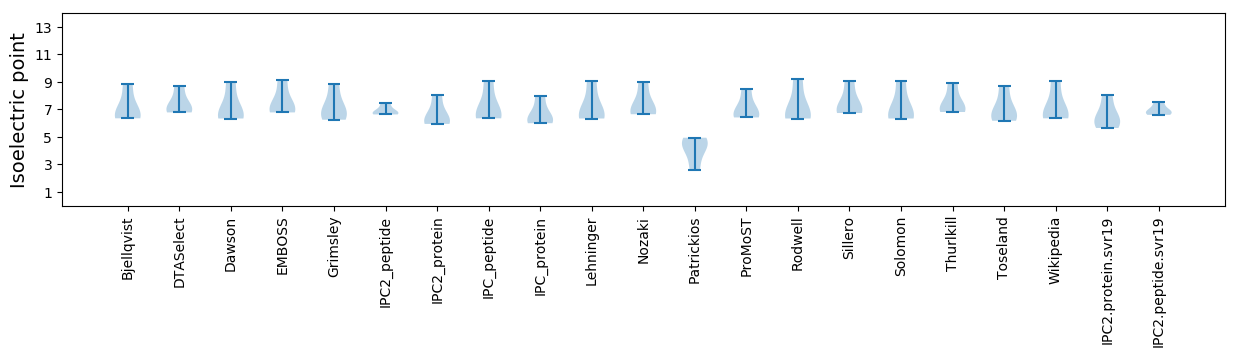
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KU60|A0A0B5KU60_9VIRU Nucleocapsid protein OS=Wuhan horsefly Virus OX=1608139 GN=N PE=4 SV=1
MM1 pKa = 7.37LKK3 pKa = 10.23KK4 pKa = 10.39YY5 pKa = 10.71LKK7 pKa = 10.32EE8 pKa = 3.78KK9 pKa = 10.38SKK11 pKa = 10.96KK12 pKa = 9.69DD13 pKa = 3.29KK14 pKa = 10.72KK15 pKa = 10.72RR16 pKa = 11.84EE17 pKa = 3.84NTLVVSTAGEE27 pKa = 4.02ASHH30 pKa = 6.8SRR32 pKa = 11.84GAIPKK37 pKa = 5.74TQRR40 pKa = 11.84IIRR43 pKa = 11.84YY44 pKa = 6.61SQRR47 pKa = 11.84EE48 pKa = 4.16LKK50 pKa = 10.34EE51 pKa = 3.46QDD53 pKa = 4.75KK54 pKa = 11.0ILQQKK59 pKa = 9.95NNGDD63 pKa = 3.84IGNMDD68 pKa = 3.81SVRR71 pKa = 11.84EE72 pKa = 4.07RR73 pKa = 11.84IFEE76 pKa = 3.82SHH78 pKa = 5.64YY79 pKa = 10.85RR80 pKa = 11.84SMEE83 pKa = 4.03TTPSEE88 pKa = 4.03LKK90 pKa = 10.99GMLEE94 pKa = 3.97LLSYY98 pKa = 9.82YY99 pKa = 9.75PRR101 pKa = 11.84APTEE105 pKa = 4.03TFTSLKK111 pKa = 9.79ISSPVQVTDD120 pKa = 3.65GLSIMEE126 pKa = 4.98IINGEE131 pKa = 4.22RR132 pKa = 11.84ISTGWKK138 pKa = 9.37HH139 pKa = 5.43PVGFVGFPRR148 pKa = 11.84IIFIFNACSPDD159 pKa = 3.69DD160 pKa = 4.25PGVIIVSLTHH170 pKa = 6.85KK171 pKa = 10.39GIKK174 pKa = 9.65DD175 pKa = 3.4PCGATLKK182 pKa = 10.67RR183 pKa = 11.84FTGPVNKK190 pKa = 8.6TWSIAFSLKK199 pKa = 10.64HH200 pKa = 5.21MVAIQDD206 pKa = 3.51IEE208 pKa = 3.97KK209 pKa = 10.59LQIHH213 pKa = 6.4CLVEE217 pKa = 3.96DD218 pKa = 3.95SNRR221 pKa = 11.84EE222 pKa = 3.62DD223 pKa = 3.74SVIGKK228 pKa = 9.88LDD230 pKa = 3.29MHH232 pKa = 6.5FTLDD236 pKa = 4.13LSTTPMIINYY246 pKa = 9.38PEE248 pKa = 3.63TTYY251 pKa = 10.87YY252 pKa = 9.74IHH254 pKa = 7.93PEE256 pKa = 3.99LDD258 pKa = 3.2VKK260 pKa = 10.99KK261 pKa = 10.54YY262 pKa = 10.38IEE264 pKa = 5.11EE265 pKa = 3.81IDD267 pKa = 3.42KK268 pKa = 11.35ASIKK272 pKa = 10.35KK273 pKa = 9.76FRR275 pKa = 11.84EE276 pKa = 3.46EE277 pKa = 4.0DD278 pKa = 3.19EE279 pKa = 4.55ARR281 pKa = 11.84LLRR284 pKa = 11.84TRR286 pKa = 11.84NYY288 pKa = 10.42AISKK292 pKa = 7.99STGVDD297 pKa = 3.44DD298 pKa = 5.72SIHH301 pKa = 5.95IQPSAPMNEE310 pKa = 3.92NEE312 pKa = 5.15KK313 pKa = 10.57IIINLDD319 pKa = 3.29PKK321 pKa = 10.64IDD323 pKa = 3.92DD324 pKa = 4.17GFNLKK329 pKa = 10.25SNKK332 pKa = 9.79GADD335 pKa = 3.79SLHH338 pKa = 5.22TTQEE342 pKa = 3.64EE343 pKa = 4.35EE344 pKa = 3.79IRR346 pKa = 11.84RR347 pKa = 11.84KK348 pKa = 10.3LRR350 pKa = 11.84DD351 pKa = 3.03LAIRR355 pKa = 11.84GKK357 pKa = 10.17KK358 pKa = 7.22QQ359 pKa = 2.74
MM1 pKa = 7.37LKK3 pKa = 10.23KK4 pKa = 10.39YY5 pKa = 10.71LKK7 pKa = 10.32EE8 pKa = 3.78KK9 pKa = 10.38SKK11 pKa = 10.96KK12 pKa = 9.69DD13 pKa = 3.29KK14 pKa = 10.72KK15 pKa = 10.72RR16 pKa = 11.84EE17 pKa = 3.84NTLVVSTAGEE27 pKa = 4.02ASHH30 pKa = 6.8SRR32 pKa = 11.84GAIPKK37 pKa = 5.74TQRR40 pKa = 11.84IIRR43 pKa = 11.84YY44 pKa = 6.61SQRR47 pKa = 11.84EE48 pKa = 4.16LKK50 pKa = 10.34EE51 pKa = 3.46QDD53 pKa = 4.75KK54 pKa = 11.0ILQQKK59 pKa = 9.95NNGDD63 pKa = 3.84IGNMDD68 pKa = 3.81SVRR71 pKa = 11.84EE72 pKa = 4.07RR73 pKa = 11.84IFEE76 pKa = 3.82SHH78 pKa = 5.64YY79 pKa = 10.85RR80 pKa = 11.84SMEE83 pKa = 4.03TTPSEE88 pKa = 4.03LKK90 pKa = 10.99GMLEE94 pKa = 3.97LLSYY98 pKa = 9.82YY99 pKa = 9.75PRR101 pKa = 11.84APTEE105 pKa = 4.03TFTSLKK111 pKa = 9.79ISSPVQVTDD120 pKa = 3.65GLSIMEE126 pKa = 4.98IINGEE131 pKa = 4.22RR132 pKa = 11.84ISTGWKK138 pKa = 9.37HH139 pKa = 5.43PVGFVGFPRR148 pKa = 11.84IIFIFNACSPDD159 pKa = 3.69DD160 pKa = 4.25PGVIIVSLTHH170 pKa = 6.85KK171 pKa = 10.39GIKK174 pKa = 9.65DD175 pKa = 3.4PCGATLKK182 pKa = 10.67RR183 pKa = 11.84FTGPVNKK190 pKa = 8.6TWSIAFSLKK199 pKa = 10.64HH200 pKa = 5.21MVAIQDD206 pKa = 3.51IEE208 pKa = 3.97KK209 pKa = 10.59LQIHH213 pKa = 6.4CLVEE217 pKa = 3.96DD218 pKa = 3.95SNRR221 pKa = 11.84EE222 pKa = 3.62DD223 pKa = 3.74SVIGKK228 pKa = 9.88LDD230 pKa = 3.29MHH232 pKa = 6.5FTLDD236 pKa = 4.13LSTTPMIINYY246 pKa = 9.38PEE248 pKa = 3.63TTYY251 pKa = 10.87YY252 pKa = 9.74IHH254 pKa = 7.93PEE256 pKa = 3.99LDD258 pKa = 3.2VKK260 pKa = 10.99KK261 pKa = 10.54YY262 pKa = 10.38IEE264 pKa = 5.11EE265 pKa = 3.81IDD267 pKa = 3.42KK268 pKa = 11.35ASIKK272 pKa = 10.35KK273 pKa = 9.76FRR275 pKa = 11.84EE276 pKa = 3.46EE277 pKa = 4.0DD278 pKa = 3.19EE279 pKa = 4.55ARR281 pKa = 11.84LLRR284 pKa = 11.84TRR286 pKa = 11.84NYY288 pKa = 10.42AISKK292 pKa = 7.99STGVDD297 pKa = 3.44DD298 pKa = 5.72SIHH301 pKa = 5.95IQPSAPMNEE310 pKa = 3.92NEE312 pKa = 5.15KK313 pKa = 10.57IIINLDD319 pKa = 3.29PKK321 pKa = 10.64IDD323 pKa = 3.92DD324 pKa = 4.17GFNLKK329 pKa = 10.25SNKK332 pKa = 9.79GADD335 pKa = 3.79SLHH338 pKa = 5.22TTQEE342 pKa = 3.64EE343 pKa = 4.35EE344 pKa = 3.79IRR346 pKa = 11.84RR347 pKa = 11.84KK348 pKa = 10.3LRR350 pKa = 11.84DD351 pKa = 3.03LAIRR355 pKa = 11.84GKK357 pKa = 10.17KK358 pKa = 7.22QQ359 pKa = 2.74
Molecular weight: 40.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4747 |
359 |
3117 |
1186.8 |
137.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.097 ± 0.448 | 1.896 ± 0.742 |
5.751 ± 0.15 | 7.183 ± 0.557 |
4.761 ± 0.28 | 3.855 ± 0.493 |
2.359 ± 0.176 | 9.164 ± 0.542 |
8.237 ± 0.312 | 9.585 ± 0.319 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.928 ± 0.336 | 5.898 ± 0.277 |
3.476 ± 0.322 | 3.16 ± 0.282 |
4.403 ± 0.393 | 8.995 ± 0.186 |
5.309 ± 0.312 | 4.803 ± 0.515 |
0.969 ± 0.088 | 4.171 ± 0.332 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
