
Actinomadura chibensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Thermomonosporaceae; Actinomadura
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
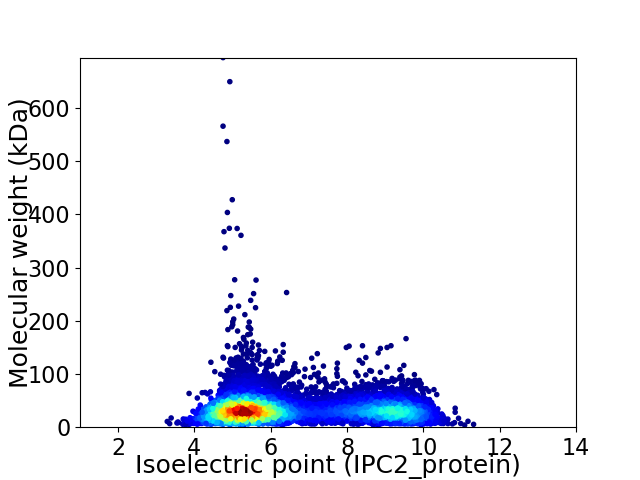
Virtual 2D-PAGE plot for 8099 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
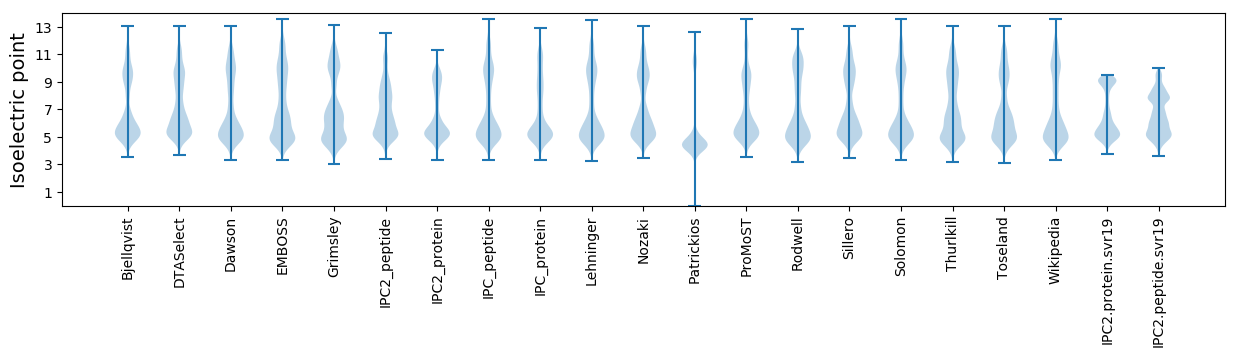
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5D0NF79|A0A5D0NF79_9ACTN Uncharacterized protein OS=Actinomadura chibensis OX=392828 GN=FXF69_29380 PE=4 SV=1
MM1 pKa = 7.24TSYY4 pKa = 10.71IVVFGLIVVVVLVVVLVALGLRR26 pKa = 11.84ASKK29 pKa = 10.57AGRR32 pKa = 11.84DD33 pKa = 3.64DD34 pKa = 5.11DD35 pKa = 5.55DD36 pKa = 4.22WMDD39 pKa = 5.38DD40 pKa = 3.53EE41 pKa = 5.13PKK43 pKa = 10.28QPRR46 pKa = 11.84GRR48 pKa = 11.84RR49 pKa = 11.84GVPPPEE55 pKa = 5.04DD56 pKa = 2.88MGGPDD61 pKa = 3.43EE62 pKa = 4.28YY63 pKa = 11.29GGYY66 pKa = 10.44GDD68 pKa = 5.91GYY70 pKa = 10.22DD71 pKa = 3.17TGYY74 pKa = 11.01DD75 pKa = 3.01AGYY78 pKa = 9.79DD79 pKa = 3.19RR80 pKa = 11.84HH81 pKa = 6.16GAPEE85 pKa = 3.94PAAAPDD91 pKa = 3.45RR92 pKa = 11.84RR93 pKa = 11.84VAGGPLAAPTSAPAPPRR110 pKa = 11.84PSAPASDD117 pKa = 3.69EE118 pKa = 4.29MEE120 pKa = 5.6DD121 pKa = 3.45DD122 pKa = 5.39DD123 pKa = 4.41YY124 pKa = 11.55WATITFDD131 pKa = 3.31KK132 pKa = 11.02PKK134 pKa = 10.46FPWQHH139 pKa = 6.58DD140 pKa = 4.15EE141 pKa = 3.84NGEE144 pKa = 4.11RR145 pKa = 11.84PDD147 pKa = 3.59VDD149 pKa = 3.73PAADD153 pKa = 4.08PLNAQAPADD162 pKa = 4.17QPPVDD167 pKa = 4.32QPGPPHH173 pKa = 7.26PSLTQPALTQPVSMNADD190 pKa = 3.65GQPLGLDD197 pKa = 3.7GQPLGMGGGTGPQQVPAPGGFQGASPFAGHH227 pKa = 7.39DD228 pKa = 3.75PGSTALDD235 pKa = 4.99PIPTDD240 pKa = 3.68LGGQGMPGGQPSYY253 pKa = 10.57GGPGVPDD260 pKa = 3.24QQVYY264 pKa = 10.27GGQEE268 pKa = 3.62PQPAYY273 pKa = 10.38GAADD277 pKa = 3.59YY278 pKa = 11.48GGGEE282 pKa = 4.16PSYY285 pKa = 11.42GGGGQDD291 pKa = 4.16QGYY294 pKa = 10.03SDD296 pKa = 3.61QPQYY300 pKa = 11.33GSHH303 pKa = 6.49EE304 pKa = 4.19PPSYY308 pKa = 11.23GDD310 pKa = 3.67YY311 pKa = 11.18SSDD314 pKa = 3.56PLGGRR319 pKa = 11.84PPASPPSGGRR329 pKa = 11.84PDD331 pKa = 4.18AFGPSTTGPSTTGPSTTGPSTTGPTPAGPGGPLGGDD367 pKa = 3.4PRR369 pKa = 11.84VSDD372 pKa = 4.28PLGLPLGGRR381 pKa = 11.84PDD383 pKa = 3.88EE384 pKa = 4.58PAVPPPAPPAPPAPAAPAAHH404 pKa = 6.55TGGGSDD410 pKa = 3.51TDD412 pKa = 3.59GHH414 pKa = 6.47KK415 pKa = 10.89LPTVDD420 pKa = 5.42EE421 pKa = 4.2LLQRR425 pKa = 11.84IQNDD429 pKa = 3.79RR430 pKa = 11.84QRR432 pKa = 11.84SSGPSGSDD440 pKa = 2.78QGGSYY445 pKa = 10.88GGGSLNDD452 pKa = 4.02PLGDD456 pKa = 3.77PLGTGSFGGSSGSAAPGGGGNTGPWPSPGQSSGYY490 pKa = 10.38DD491 pKa = 3.12SGGGSGLPSGSGYY504 pKa = 10.8DD505 pKa = 3.47SGLGAPSGYY514 pKa = 10.72GQGQQNDD521 pKa = 5.03GYY523 pKa = 8.33PTAPAYY529 pKa = 10.51GEE531 pKa = 4.35TASPRR536 pKa = 11.84YY537 pKa = 9.51DD538 pKa = 3.69DD539 pKa = 4.56PLSGGRR545 pKa = 11.84EE546 pKa = 4.37SFGGTYY552 pKa = 10.48GGGAEE557 pKa = 4.35GGSNGGSSGSGGSGGVYY574 pKa = 10.6GDD576 pKa = 4.58FSGSSYY582 pKa = 11.54NGGNDD587 pKa = 3.92PLSAPPDD594 pKa = 3.94PNATQAYY601 pKa = 8.12GQGGYY606 pKa = 9.98SGGYY610 pKa = 7.06QQGGYY615 pKa = 7.39QQGQQNDD622 pKa = 3.62GSQYY626 pKa = 10.77GSRR629 pKa = 11.84PPADD633 pKa = 2.97DD634 pKa = 3.26WEE636 pKa = 4.34NYY638 pKa = 8.27RR639 pKa = 11.84RR640 pKa = 3.78
MM1 pKa = 7.24TSYY4 pKa = 10.71IVVFGLIVVVVLVVVLVALGLRR26 pKa = 11.84ASKK29 pKa = 10.57AGRR32 pKa = 11.84DD33 pKa = 3.64DD34 pKa = 5.11DD35 pKa = 5.55DD36 pKa = 4.22WMDD39 pKa = 5.38DD40 pKa = 3.53EE41 pKa = 5.13PKK43 pKa = 10.28QPRR46 pKa = 11.84GRR48 pKa = 11.84RR49 pKa = 11.84GVPPPEE55 pKa = 5.04DD56 pKa = 2.88MGGPDD61 pKa = 3.43EE62 pKa = 4.28YY63 pKa = 11.29GGYY66 pKa = 10.44GDD68 pKa = 5.91GYY70 pKa = 10.22DD71 pKa = 3.17TGYY74 pKa = 11.01DD75 pKa = 3.01AGYY78 pKa = 9.79DD79 pKa = 3.19RR80 pKa = 11.84HH81 pKa = 6.16GAPEE85 pKa = 3.94PAAAPDD91 pKa = 3.45RR92 pKa = 11.84RR93 pKa = 11.84VAGGPLAAPTSAPAPPRR110 pKa = 11.84PSAPASDD117 pKa = 3.69EE118 pKa = 4.29MEE120 pKa = 5.6DD121 pKa = 3.45DD122 pKa = 5.39DD123 pKa = 4.41YY124 pKa = 11.55WATITFDD131 pKa = 3.31KK132 pKa = 11.02PKK134 pKa = 10.46FPWQHH139 pKa = 6.58DD140 pKa = 4.15EE141 pKa = 3.84NGEE144 pKa = 4.11RR145 pKa = 11.84PDD147 pKa = 3.59VDD149 pKa = 3.73PAADD153 pKa = 4.08PLNAQAPADD162 pKa = 4.17QPPVDD167 pKa = 4.32QPGPPHH173 pKa = 7.26PSLTQPALTQPVSMNADD190 pKa = 3.65GQPLGLDD197 pKa = 3.7GQPLGMGGGTGPQQVPAPGGFQGASPFAGHH227 pKa = 7.39DD228 pKa = 3.75PGSTALDD235 pKa = 4.99PIPTDD240 pKa = 3.68LGGQGMPGGQPSYY253 pKa = 10.57GGPGVPDD260 pKa = 3.24QQVYY264 pKa = 10.27GGQEE268 pKa = 3.62PQPAYY273 pKa = 10.38GAADD277 pKa = 3.59YY278 pKa = 11.48GGGEE282 pKa = 4.16PSYY285 pKa = 11.42GGGGQDD291 pKa = 4.16QGYY294 pKa = 10.03SDD296 pKa = 3.61QPQYY300 pKa = 11.33GSHH303 pKa = 6.49EE304 pKa = 4.19PPSYY308 pKa = 11.23GDD310 pKa = 3.67YY311 pKa = 11.18SSDD314 pKa = 3.56PLGGRR319 pKa = 11.84PPASPPSGGRR329 pKa = 11.84PDD331 pKa = 4.18AFGPSTTGPSTTGPSTTGPSTTGPTPAGPGGPLGGDD367 pKa = 3.4PRR369 pKa = 11.84VSDD372 pKa = 4.28PLGLPLGGRR381 pKa = 11.84PDD383 pKa = 3.88EE384 pKa = 4.58PAVPPPAPPAPPAPAAPAAHH404 pKa = 6.55TGGGSDD410 pKa = 3.51TDD412 pKa = 3.59GHH414 pKa = 6.47KK415 pKa = 10.89LPTVDD420 pKa = 5.42EE421 pKa = 4.2LLQRR425 pKa = 11.84IQNDD429 pKa = 3.79RR430 pKa = 11.84QRR432 pKa = 11.84SSGPSGSDD440 pKa = 2.78QGGSYY445 pKa = 10.88GGGSLNDD452 pKa = 4.02PLGDD456 pKa = 3.77PLGTGSFGGSSGSAAPGGGGNTGPWPSPGQSSGYY490 pKa = 10.38DD491 pKa = 3.12SGGGSGLPSGSGYY504 pKa = 10.8DD505 pKa = 3.47SGLGAPSGYY514 pKa = 10.72GQGQQNDD521 pKa = 5.03GYY523 pKa = 8.33PTAPAYY529 pKa = 10.51GEE531 pKa = 4.35TASPRR536 pKa = 11.84YY537 pKa = 9.51DD538 pKa = 3.69DD539 pKa = 4.56PLSGGRR545 pKa = 11.84EE546 pKa = 4.37SFGGTYY552 pKa = 10.48GGGAEE557 pKa = 4.35GGSNGGSSGSGGSGGVYY574 pKa = 10.6GDD576 pKa = 4.58FSGSSYY582 pKa = 11.54NGGNDD587 pKa = 3.92PLSAPPDD594 pKa = 3.94PNATQAYY601 pKa = 8.12GQGGYY606 pKa = 9.98SGGYY610 pKa = 7.06QQGGYY615 pKa = 7.39QQGQQNDD622 pKa = 3.62GSQYY626 pKa = 10.77GSRR629 pKa = 11.84PPADD633 pKa = 2.97DD634 pKa = 3.26WEE636 pKa = 4.34NYY638 pKa = 8.27RR639 pKa = 11.84RR640 pKa = 3.78
Molecular weight: 63.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5D0NYS9|A0A5D0NYS9_9ACTN DUF3427 domain-containing protein OS=Actinomadura chibensis OX=392828 GN=FXF69_11965 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 5.34KK15 pKa = 10.47KK16 pKa = 8.84HH17 pKa = 5.5GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILANRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.24GRR40 pKa = 11.84ARR42 pKa = 11.84IAVV45 pKa = 3.5
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 5.34KK15 pKa = 10.47KK16 pKa = 8.84HH17 pKa = 5.5GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILANRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.24GRR40 pKa = 11.84ARR42 pKa = 11.84IAVV45 pKa = 3.5
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2744976 |
27 |
6628 |
338.9 |
36.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.08 ± 0.039 | 0.786 ± 0.008 |
6.21 ± 0.022 | 5.525 ± 0.026 |
2.776 ± 0.014 | 9.755 ± 0.026 |
2.156 ± 0.012 | 3.161 ± 0.017 |
1.923 ± 0.021 | 10.292 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.761 ± 0.01 | 1.672 ± 0.012 |
6.361 ± 0.026 | 2.337 ± 0.014 |
8.829 ± 0.026 | 4.68 ± 0.017 |
5.515 ± 0.023 | 8.701 ± 0.028 |
1.488 ± 0.009 | 1.992 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
