
Scylla serrata reovirus SZ-2007
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; unclassified Reoviridae
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
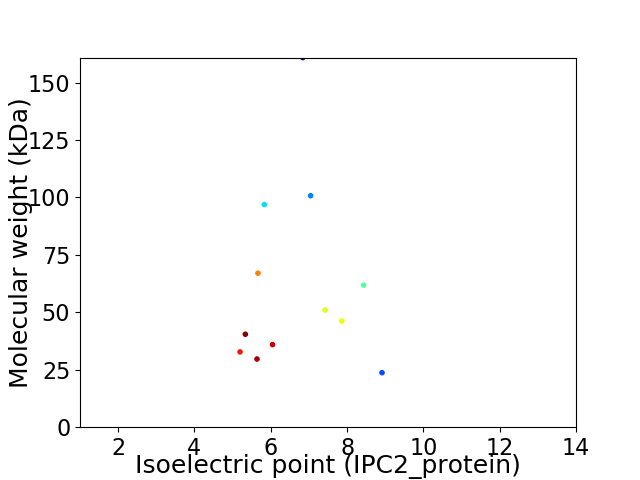
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
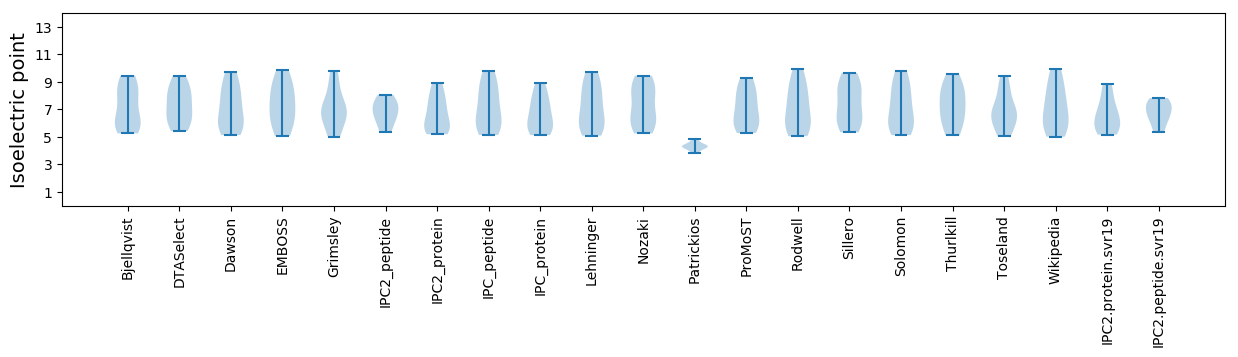
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G9BDA7|G9BDA7_9REOV VP11 OS=Scylla serrata reovirus SZ-2007 OX=458682 GN=MCRV_s11gp1 PE=4 SV=1
MM1 pKa = 7.86ADD3 pKa = 3.84SITGRR8 pKa = 11.84SGAGLPMSHH17 pKa = 6.78TYY19 pKa = 10.58GYY21 pKa = 10.73QILLQKK27 pKa = 9.53QQYY30 pKa = 10.23DD31 pKa = 3.19HH32 pKa = 7.31DD33 pKa = 4.21TTVPSEE39 pKa = 4.45STWTPFEE46 pKa = 5.29SNDD49 pKa = 3.22QYY51 pKa = 11.52TIARR55 pKa = 11.84DD56 pKa = 3.47TEE58 pKa = 4.23AYY60 pKa = 10.98LNFFQIGTRR69 pKa = 11.84VFFTVTPGPYY79 pKa = 10.26GDD81 pKa = 4.15LQEE84 pKa = 4.26QFRR87 pKa = 11.84SLRR90 pKa = 11.84LKK92 pKa = 10.18VIWTKK97 pKa = 11.11NGGLAYY103 pKa = 10.3SEE105 pKa = 4.28EE106 pKa = 4.42RR107 pKa = 11.84TSILHH112 pKa = 5.71WLQRR116 pKa = 11.84LANTKK121 pKa = 10.42SILTQCPVFLVCDD134 pKa = 3.69NFEE137 pKa = 4.09RR138 pKa = 11.84LYY140 pKa = 11.46VLLPCTMNKK149 pKa = 10.17DD150 pKa = 3.85YY151 pKa = 10.89PCGSFAVRR159 pKa = 11.84KK160 pKa = 9.65KK161 pKa = 10.28IPKK164 pKa = 9.54QYY166 pKa = 11.18AVLARR171 pKa = 11.84LMNLVPSSKK180 pKa = 10.32QEE182 pKa = 3.87NNLYY186 pKa = 9.65VYY188 pKa = 11.41NMLKK192 pKa = 10.5ALRR195 pKa = 11.84LRR197 pKa = 11.84SDD199 pKa = 4.1DD200 pKa = 3.69STEE203 pKa = 5.45LIPTDD208 pKa = 3.37DD209 pKa = 4.43AARR212 pKa = 11.84VMAPYY217 pKa = 9.08LTDD220 pKa = 3.77EE221 pKa = 4.75DD222 pKa = 4.04TSSIIRR228 pKa = 11.84GALLGLEE235 pKa = 4.48PDD237 pKa = 4.01SIVIRR242 pKa = 11.84WCNLLTDD249 pKa = 4.09YY250 pKa = 11.02DD251 pKa = 4.06IEE253 pKa = 4.41LTTSQWYY260 pKa = 10.12FDD262 pKa = 3.4TKK264 pKa = 11.06VIVEE268 pKa = 4.65KK269 pKa = 9.72MWKK272 pKa = 7.33TQSTPKK278 pKa = 9.84IDD280 pKa = 3.44ASTFLPEE287 pKa = 3.86YY288 pKa = 11.36DD289 pKa = 3.8MLNLLSIKK297 pKa = 9.94KK298 pKa = 10.23GKK300 pKa = 9.87ALYY303 pKa = 9.41EE304 pKa = 4.1DD305 pKa = 3.92WVASAKK311 pKa = 10.63EE312 pKa = 4.09SDD314 pKa = 3.61HH315 pKa = 6.91NGALVRR321 pKa = 11.84AGKK324 pKa = 10.51YY325 pKa = 10.38GIEE328 pKa = 4.19NGDD331 pKa = 3.28LTGRR335 pKa = 11.84EE336 pKa = 3.81TFIPVWLEE344 pKa = 3.6QLITKK349 pKa = 10.28ADD351 pKa = 3.24VYY353 pKa = 10.9YY354 pKa = 11.25
MM1 pKa = 7.86ADD3 pKa = 3.84SITGRR8 pKa = 11.84SGAGLPMSHH17 pKa = 6.78TYY19 pKa = 10.58GYY21 pKa = 10.73QILLQKK27 pKa = 9.53QQYY30 pKa = 10.23DD31 pKa = 3.19HH32 pKa = 7.31DD33 pKa = 4.21TTVPSEE39 pKa = 4.45STWTPFEE46 pKa = 5.29SNDD49 pKa = 3.22QYY51 pKa = 11.52TIARR55 pKa = 11.84DD56 pKa = 3.47TEE58 pKa = 4.23AYY60 pKa = 10.98LNFFQIGTRR69 pKa = 11.84VFFTVTPGPYY79 pKa = 10.26GDD81 pKa = 4.15LQEE84 pKa = 4.26QFRR87 pKa = 11.84SLRR90 pKa = 11.84LKK92 pKa = 10.18VIWTKK97 pKa = 11.11NGGLAYY103 pKa = 10.3SEE105 pKa = 4.28EE106 pKa = 4.42RR107 pKa = 11.84TSILHH112 pKa = 5.71WLQRR116 pKa = 11.84LANTKK121 pKa = 10.42SILTQCPVFLVCDD134 pKa = 3.69NFEE137 pKa = 4.09RR138 pKa = 11.84LYY140 pKa = 11.46VLLPCTMNKK149 pKa = 10.17DD150 pKa = 3.85YY151 pKa = 10.89PCGSFAVRR159 pKa = 11.84KK160 pKa = 9.65KK161 pKa = 10.28IPKK164 pKa = 9.54QYY166 pKa = 11.18AVLARR171 pKa = 11.84LMNLVPSSKK180 pKa = 10.32QEE182 pKa = 3.87NNLYY186 pKa = 9.65VYY188 pKa = 11.41NMLKK192 pKa = 10.5ALRR195 pKa = 11.84LRR197 pKa = 11.84SDD199 pKa = 4.1DD200 pKa = 3.69STEE203 pKa = 5.45LIPTDD208 pKa = 3.37DD209 pKa = 4.43AARR212 pKa = 11.84VMAPYY217 pKa = 9.08LTDD220 pKa = 3.77EE221 pKa = 4.75DD222 pKa = 4.04TSSIIRR228 pKa = 11.84GALLGLEE235 pKa = 4.48PDD237 pKa = 4.01SIVIRR242 pKa = 11.84WCNLLTDD249 pKa = 4.09YY250 pKa = 11.02DD251 pKa = 4.06IEE253 pKa = 4.41LTTSQWYY260 pKa = 10.12FDD262 pKa = 3.4TKK264 pKa = 11.06VIVEE268 pKa = 4.65KK269 pKa = 9.72MWKK272 pKa = 7.33TQSTPKK278 pKa = 9.84IDD280 pKa = 3.44ASTFLPEE287 pKa = 3.86YY288 pKa = 11.36DD289 pKa = 3.8MLNLLSIKK297 pKa = 9.94KK298 pKa = 10.23GKK300 pKa = 9.87ALYY303 pKa = 9.41EE304 pKa = 4.1DD305 pKa = 3.92WVASAKK311 pKa = 10.63EE312 pKa = 4.09SDD314 pKa = 3.61HH315 pKa = 6.91NGALVRR321 pKa = 11.84AGKK324 pKa = 10.51YY325 pKa = 10.38GIEE328 pKa = 4.19NGDD331 pKa = 3.28LTGRR335 pKa = 11.84EE336 pKa = 3.81TFIPVWLEE344 pKa = 3.6QLITKK349 pKa = 10.28ADD351 pKa = 3.24VYY353 pKa = 10.9YY354 pKa = 11.25
Molecular weight: 40.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H9NEU8|H9NEU8_9REOV Uncharacterized protein OS=Scylla serrata reovirus SZ-2007 OX=458682 PE=4 SV=1
MM1 pKa = 7.19NWSKK5 pKa = 11.16AINFQPFMLEE15 pKa = 3.66TRR17 pKa = 11.84PPLTTIPIMDD27 pKa = 3.77QLVEE31 pKa = 3.76IGEE34 pKa = 4.33RR35 pKa = 11.84SNQKK39 pKa = 9.57WSMTDD44 pKa = 2.76RR45 pKa = 11.84LFFAIRR51 pKa = 11.84KK52 pKa = 8.0INPIFVTSSQIPSKK66 pKa = 10.63FDD68 pKa = 3.24YY69 pKa = 10.54TILQMPTQLIASLKK83 pKa = 8.12EE84 pKa = 3.91TLLFLAFSYY93 pKa = 10.12YY94 pKa = 10.27LRR96 pKa = 11.84EE97 pKa = 4.14YY98 pKa = 9.45QDD100 pKa = 4.12KK101 pKa = 9.27VGQMKK106 pKa = 9.36FYY108 pKa = 9.98PVAMKK113 pKa = 11.03NMIPIVNYY121 pKa = 10.72LKK123 pKa = 10.97DD124 pKa = 3.5RR125 pKa = 11.84VHH127 pKa = 6.76NNFDD131 pKa = 3.51TTLEE135 pKa = 3.7QAYY138 pKa = 9.12RR139 pKa = 11.84QNVVHH144 pKa = 6.41TLSASDD150 pKa = 4.88AFDD153 pKa = 4.49LLSGMIATTRR163 pKa = 11.84LDD165 pKa = 4.49LIQRR169 pKa = 11.84TRR171 pKa = 11.84ICPEE175 pKa = 3.85LLNVLNKK182 pKa = 9.85MSFILIYY189 pKa = 10.61APNRR193 pKa = 11.84PSILSWKK200 pKa = 8.75NQSS203 pKa = 2.99
MM1 pKa = 7.19NWSKK5 pKa = 11.16AINFQPFMLEE15 pKa = 3.66TRR17 pKa = 11.84PPLTTIPIMDD27 pKa = 3.77QLVEE31 pKa = 3.76IGEE34 pKa = 4.33RR35 pKa = 11.84SNQKK39 pKa = 9.57WSMTDD44 pKa = 2.76RR45 pKa = 11.84LFFAIRR51 pKa = 11.84KK52 pKa = 8.0INPIFVTSSQIPSKK66 pKa = 10.63FDD68 pKa = 3.24YY69 pKa = 10.54TILQMPTQLIASLKK83 pKa = 8.12EE84 pKa = 3.91TLLFLAFSYY93 pKa = 10.12YY94 pKa = 10.27LRR96 pKa = 11.84EE97 pKa = 4.14YY98 pKa = 9.45QDD100 pKa = 4.12KK101 pKa = 9.27VGQMKK106 pKa = 9.36FYY108 pKa = 9.98PVAMKK113 pKa = 11.03NMIPIVNYY121 pKa = 10.72LKK123 pKa = 10.97DD124 pKa = 3.5RR125 pKa = 11.84VHH127 pKa = 6.76NNFDD131 pKa = 3.51TTLEE135 pKa = 3.7QAYY138 pKa = 9.12RR139 pKa = 11.84QNVVHH144 pKa = 6.41TLSASDD150 pKa = 4.88AFDD153 pKa = 4.49LLSGMIATTRR163 pKa = 11.84LDD165 pKa = 4.49LIQRR169 pKa = 11.84TRR171 pKa = 11.84ICPEE175 pKa = 3.85LLNVLNKK182 pKa = 9.85MSFILIYY189 pKa = 10.61APNRR193 pKa = 11.84PSILSWKK200 pKa = 8.75NQSS203 pKa = 2.99
Molecular weight: 23.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6608 |
203 |
1422 |
550.7 |
62.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.568 ± 0.615 | 0.878 ± 0.199 |
6.326 ± 0.259 | 5.16 ± 0.183 |
3.435 ± 0.189 | 5.145 ± 0.409 |
2.467 ± 0.298 | 7.234 ± 0.386 |
5.176 ± 0.279 | 9.11 ± 0.331 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.906 ± 0.221 | 4.54 ± 0.206 |
4.222 ± 0.395 | 4.54 ± 0.098 |
6.129 ± 0.393 | 7.491 ± 0.262 |
6.674 ± 0.479 | 7.052 ± 0.368 |
0.938 ± 0.138 | 4.01 ± 0.285 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
