
Geodermatophilus obscurus (strain ATCC 25078 / DSM 43160 / JCM 3152 / KCC A-0152 / KCTC 9177 / NBRC 13315 / NRRL B-3577 / G-20)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Geodermatophilus; Geodermatophilus obscurus
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
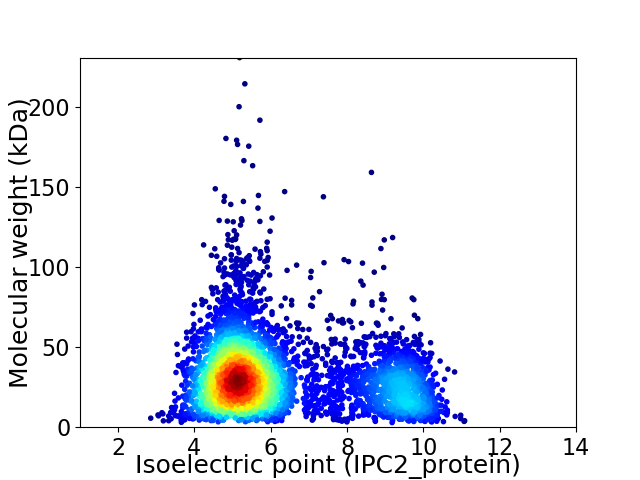
Virtual 2D-PAGE plot for 4795 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D2S9Z7|D2S9Z7_GEOOG Fibronectin type III domain protein OS=Geodermatophilus obscurus (strain ATCC 25078 / DSM 43160 / JCM 3152 / KCC A-0152 / KCTC 9177 / NBRC 13315 / NRRL B-3577 / G-20) OX=526225 GN=Gobs_1101 PE=4 SV=1
MM1 pKa = 7.65RR2 pKa = 11.84LSKK5 pKa = 10.67RR6 pKa = 11.84SAALIATGLTGAMVLSACGGGGDD29 pKa = 3.92EE30 pKa = 5.6DD31 pKa = 4.45GGSGAAAADD40 pKa = 3.76GGSFTVYY47 pKa = 9.78IAEE50 pKa = 4.68PEE52 pKa = 4.08NPLIPGQTTEE62 pKa = 4.15TEE64 pKa = 4.57GAQVLYY70 pKa = 11.03SLFTGLVQYY79 pKa = 10.98DD80 pKa = 4.01SEE82 pKa = 4.64TNEE85 pKa = 3.64AVYY88 pKa = 9.94TGVADD93 pKa = 4.37SIEE96 pKa = 4.47SADD99 pKa = 3.52QTTWTVKK106 pKa = 10.78LKK108 pKa = 10.87DD109 pKa = 2.95GWTFHH114 pKa = 7.35DD115 pKa = 5.19GSPVRR120 pKa = 11.84AQSFVDD126 pKa = 3.4AWNWTAYY133 pKa = 10.39SPNAANSSYY142 pKa = 10.65FFANIAGYY150 pKa = 9.73DD151 pKa = 3.68QLQAPTDD158 pKa = 3.63DD159 pKa = 4.32AGNVTGDD166 pKa = 3.69PAATAMSGLRR176 pKa = 11.84VVDD179 pKa = 4.26DD180 pKa = 3.66LTFEE184 pKa = 4.44VTLSAPYY191 pKa = 9.76AQWPTTVGYY200 pKa = 10.17SAFYY204 pKa = 9.33PLPPAFFEE212 pKa = 4.69DD213 pKa = 3.42SAAFGEE219 pKa = 4.34QPIGNGPFRR228 pKa = 11.84ADD230 pKa = 3.26EE231 pKa = 4.33PFVPGTGVTLTRR243 pKa = 11.84YY244 pKa = 10.21DD245 pKa = 3.73EE246 pKa = 4.38YY247 pKa = 11.68GGDD250 pKa = 3.66KK251 pKa = 9.67PANAASVQYY260 pKa = 10.76VVYY263 pKa = 10.67AEE265 pKa = 3.97QATAYY270 pKa = 10.36RR271 pKa = 11.84DD272 pKa = 3.74LQAGNLDD279 pKa = 4.05IMDD282 pKa = 5.12EE283 pKa = 4.39LPPDD287 pKa = 3.82ALASAEE293 pKa = 4.2AEE295 pKa = 4.05LGDD298 pKa = 4.44RR299 pKa = 11.84LLQVEE304 pKa = 4.3QGDD307 pKa = 3.86ITSLGFPTYY316 pKa = 10.13DD317 pKa = 4.1EE318 pKa = 5.18RR319 pKa = 11.84FADD322 pKa = 3.82PNVRR326 pKa = 11.84RR327 pKa = 11.84AFSMAIDD334 pKa = 3.57RR335 pKa = 11.84EE336 pKa = 4.65SITEE340 pKa = 4.53AIFQGTRR347 pKa = 11.84IPATSFINPVVDD359 pKa = 4.82GYY361 pKa = 11.45RR362 pKa = 11.84EE363 pKa = 4.22GACDD367 pKa = 3.42VCEE370 pKa = 4.65LNVEE374 pKa = 4.19EE375 pKa = 5.2ANRR378 pKa = 11.84LLDD381 pKa = 3.33EE382 pKa = 5.09AGFDD386 pKa = 3.37RR387 pKa = 11.84SQPVDD392 pKa = 2.84LWFNAGAGHH401 pKa = 6.92EE402 pKa = 4.04IWMEE406 pKa = 3.69AVGNQIRR413 pKa = 11.84EE414 pKa = 4.31GLGVDD419 pKa = 3.46YY420 pKa = 10.1TLQGNLDD427 pKa = 3.56FAEE430 pKa = 4.18YY431 pKa = 10.76LPLQDD436 pKa = 4.0EE437 pKa = 4.51QGMTGPFRR445 pKa = 11.84SGWIMDD451 pKa = 3.65YY452 pKa = 10.74PVAEE456 pKa = 4.34NFLGPLYY463 pKa = 10.64SSTALPPGGSNVTFYY478 pKa = 11.58SNPEE482 pKa = 3.45FDD484 pKa = 5.86ALLQQGNSADD494 pKa = 3.83TNEE497 pKa = 4.42AAIQAYY503 pKa = 8.69QAAEE507 pKa = 4.03DD508 pKa = 3.9LLLRR512 pKa = 11.84DD513 pKa = 4.21MPATPLFYY521 pKa = 10.32RR522 pKa = 11.84VNQSAHH528 pKa = 5.97SEE530 pKa = 4.13NVDD533 pKa = 3.45NVVVDD538 pKa = 3.69AFNRR542 pKa = 11.84IDD544 pKa = 3.36TAAVQVVGG552 pKa = 4.19
MM1 pKa = 7.65RR2 pKa = 11.84LSKK5 pKa = 10.67RR6 pKa = 11.84SAALIATGLTGAMVLSACGGGGDD29 pKa = 3.92EE30 pKa = 5.6DD31 pKa = 4.45GGSGAAAADD40 pKa = 3.76GGSFTVYY47 pKa = 9.78IAEE50 pKa = 4.68PEE52 pKa = 4.08NPLIPGQTTEE62 pKa = 4.15TEE64 pKa = 4.57GAQVLYY70 pKa = 11.03SLFTGLVQYY79 pKa = 10.98DD80 pKa = 4.01SEE82 pKa = 4.64TNEE85 pKa = 3.64AVYY88 pKa = 9.94TGVADD93 pKa = 4.37SIEE96 pKa = 4.47SADD99 pKa = 3.52QTTWTVKK106 pKa = 10.78LKK108 pKa = 10.87DD109 pKa = 2.95GWTFHH114 pKa = 7.35DD115 pKa = 5.19GSPVRR120 pKa = 11.84AQSFVDD126 pKa = 3.4AWNWTAYY133 pKa = 10.39SPNAANSSYY142 pKa = 10.65FFANIAGYY150 pKa = 9.73DD151 pKa = 3.68QLQAPTDD158 pKa = 3.63DD159 pKa = 4.32AGNVTGDD166 pKa = 3.69PAATAMSGLRR176 pKa = 11.84VVDD179 pKa = 4.26DD180 pKa = 3.66LTFEE184 pKa = 4.44VTLSAPYY191 pKa = 9.76AQWPTTVGYY200 pKa = 10.17SAFYY204 pKa = 9.33PLPPAFFEE212 pKa = 4.69DD213 pKa = 3.42SAAFGEE219 pKa = 4.34QPIGNGPFRR228 pKa = 11.84ADD230 pKa = 3.26EE231 pKa = 4.33PFVPGTGVTLTRR243 pKa = 11.84YY244 pKa = 10.21DD245 pKa = 3.73EE246 pKa = 4.38YY247 pKa = 11.68GGDD250 pKa = 3.66KK251 pKa = 9.67PANAASVQYY260 pKa = 10.76VVYY263 pKa = 10.67AEE265 pKa = 3.97QATAYY270 pKa = 10.36RR271 pKa = 11.84DD272 pKa = 3.74LQAGNLDD279 pKa = 4.05IMDD282 pKa = 5.12EE283 pKa = 4.39LPPDD287 pKa = 3.82ALASAEE293 pKa = 4.2AEE295 pKa = 4.05LGDD298 pKa = 4.44RR299 pKa = 11.84LLQVEE304 pKa = 4.3QGDD307 pKa = 3.86ITSLGFPTYY316 pKa = 10.13DD317 pKa = 4.1EE318 pKa = 5.18RR319 pKa = 11.84FADD322 pKa = 3.82PNVRR326 pKa = 11.84RR327 pKa = 11.84AFSMAIDD334 pKa = 3.57RR335 pKa = 11.84EE336 pKa = 4.65SITEE340 pKa = 4.53AIFQGTRR347 pKa = 11.84IPATSFINPVVDD359 pKa = 4.82GYY361 pKa = 11.45RR362 pKa = 11.84EE363 pKa = 4.22GACDD367 pKa = 3.42VCEE370 pKa = 4.65LNVEE374 pKa = 4.19EE375 pKa = 5.2ANRR378 pKa = 11.84LLDD381 pKa = 3.33EE382 pKa = 5.09AGFDD386 pKa = 3.37RR387 pKa = 11.84SQPVDD392 pKa = 2.84LWFNAGAGHH401 pKa = 6.92EE402 pKa = 4.04IWMEE406 pKa = 3.69AVGNQIRR413 pKa = 11.84EE414 pKa = 4.31GLGVDD419 pKa = 3.46YY420 pKa = 10.1TLQGNLDD427 pKa = 3.56FAEE430 pKa = 4.18YY431 pKa = 10.76LPLQDD436 pKa = 4.0EE437 pKa = 4.51QGMTGPFRR445 pKa = 11.84SGWIMDD451 pKa = 3.65YY452 pKa = 10.74PVAEE456 pKa = 4.34NFLGPLYY463 pKa = 10.64SSTALPPGGSNVTFYY478 pKa = 11.58SNPEE482 pKa = 3.45FDD484 pKa = 5.86ALLQQGNSADD494 pKa = 3.83TNEE497 pKa = 4.42AAIQAYY503 pKa = 8.69QAAEE507 pKa = 4.03DD508 pKa = 3.9LLLRR512 pKa = 11.84DD513 pKa = 4.21MPATPLFYY521 pKa = 10.32RR522 pKa = 11.84VNQSAHH528 pKa = 5.97SEE530 pKa = 4.13NVDD533 pKa = 3.45NVVVDD538 pKa = 3.69AFNRR542 pKa = 11.84IDD544 pKa = 3.36TAAVQVVGG552 pKa = 4.19
Molecular weight: 59.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D2SDM7|D2SDM7_GEOOG Uncharacterized protein OS=Geodermatophilus obscurus (strain ATCC 25078 / DSM 43160 / JCM 3152 / KCC A-0152 / KCTC 9177 / NBRC 13315 / NRRL B-3577 / G-20) OX=526225 GN=Gobs_1770 PE=4 SV=1
MM1 pKa = 7.24SRR3 pKa = 11.84RR4 pKa = 11.84HH5 pKa = 5.4RR6 pKa = 11.84VLRR9 pKa = 11.84SAIAAAAAAAAGLLTRR25 pKa = 11.84SMVRR29 pKa = 11.84GVRR32 pKa = 11.84RR33 pKa = 11.84PP34 pKa = 3.11
MM1 pKa = 7.24SRR3 pKa = 11.84RR4 pKa = 11.84HH5 pKa = 5.4RR6 pKa = 11.84VLRR9 pKa = 11.84SAIAAAAAAAAGLLTRR25 pKa = 11.84SMVRR29 pKa = 11.84GVRR32 pKa = 11.84RR33 pKa = 11.84PP34 pKa = 3.11
Molecular weight: 3.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1492083 |
30 |
2124 |
311.2 |
33.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.337 ± 0.053 | 0.695 ± 0.01 |
6.228 ± 0.027 | 5.591 ± 0.03 |
2.503 ± 0.02 | 9.642 ± 0.034 |
2.063 ± 0.016 | 2.572 ± 0.025 |
1.252 ± 0.022 | 10.751 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.543 ± 0.013 | 1.386 ± 0.015 |
6.395 ± 0.031 | 2.718 ± 0.019 |
8.492 ± 0.038 | 4.727 ± 0.023 |
5.987 ± 0.026 | 9.896 ± 0.04 |
1.483 ± 0.014 | 1.739 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
