
Leptospirillum ferrooxidans (strain C2-3)
Taxonomy: cellular organisms; Bacteria; Nitrospirae; Nitrospira; Nitrospirales; Nitrospiraceae; Leptospirillum; Leptospirillum sp. Group I; Leptospirillum ferrooxidans
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
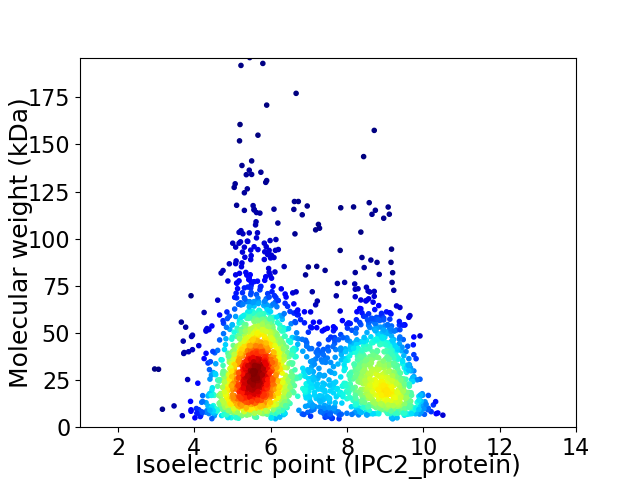
Virtual 2D-PAGE plot for 2413 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I0IQV0|I0IQV0_LEPFC Putative phage shock protein A OS=Leptospirillum ferrooxidans (strain C2-3) OX=1162668 GN=pspA PE=3 SV=1
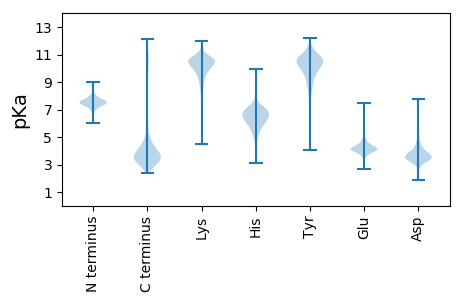
MM1 pKa = 7.55HH2 pKa = 7.96LFPMVILSASLLMASCKK19 pKa = 10.43ASNLDD24 pKa = 3.81LSSNGQAGALTSAPTQTATVSGTAVDD50 pKa = 4.12GFIDD54 pKa = 3.36QGEE57 pKa = 4.72VVLSTFPPGAAVLGSATTDD76 pKa = 2.95TSGRR80 pKa = 11.84FSIQTTPIDD89 pKa = 3.66SAALYY94 pKa = 10.4QLTLTGGSSIDD105 pKa = 3.59FASGYY110 pKa = 10.26SISLSSLDD118 pKa = 3.66SLSTIVTGKK127 pKa = 8.12TLLSSPVTITIMTTLMANQAQGMINRR153 pKa = 11.84GAAPLASFQNALGLWSGLLGFDD175 pKa = 3.71PTTTPVTDD183 pKa = 3.77PTAGPEE189 pKa = 4.28SMGYY193 pKa = 8.07PAIYY197 pKa = 10.45GLILAGFSQMAMAIGVNDD215 pKa = 4.1HH216 pKa = 7.0LSPGTSNTLGLLSVLNSDD234 pKa = 3.83ISDD237 pKa = 3.72GVLDD241 pKa = 4.04GLKK244 pKa = 10.59PGDD247 pKa = 4.41PNPLTYY253 pKa = 9.53YY254 pKa = 10.29TYY256 pKa = 10.99TLTADD261 pKa = 3.83TLRR264 pKa = 11.84KK265 pKa = 10.19DD266 pKa = 3.41IADD269 pKa = 3.64STLEE273 pKa = 3.94FLWGDD278 pKa = 3.81RR279 pKa = 11.84NQSGLAPIDD288 pKa = 3.37VSGSVNALALDD299 pKa = 3.92SSSLFPGNPLPTLPDD314 pKa = 4.31PGSPLITVSSPVNNAYY330 pKa = 10.77YY331 pKa = 11.01NNTLPLVATASDD343 pKa = 3.5ALGIGLFSVTSTTLPLPVNPPGSTSIQTGVDD374 pKa = 2.76ILGIPDD380 pKa = 4.02GPLTTVFSAEE390 pKa = 3.94NLAGTVSTQTISLSIDD406 pKa = 3.02NTAPTLQNLSPISATSYY423 pKa = 10.29DD424 pKa = 3.18ICGSTNYY431 pKa = 9.87SVQVTGQMVDD441 pKa = 3.19TGSGPSRR448 pKa = 11.84IQVQEE453 pKa = 3.98TAPVSQAISAYY464 pKa = 7.81STVVNRR470 pKa = 11.84TTSNVSFYY478 pKa = 11.53VFAPVNSPCKK488 pKa = 10.28AVTFTYY494 pKa = 10.79NITVYY499 pKa = 10.71DD500 pKa = 3.78ALNNQRR506 pKa = 11.84TIPYY510 pKa = 10.74SMIIANN516 pKa = 3.85
MM1 pKa = 7.55HH2 pKa = 7.96LFPMVILSASLLMASCKK19 pKa = 10.43ASNLDD24 pKa = 3.81LSSNGQAGALTSAPTQTATVSGTAVDD50 pKa = 4.12GFIDD54 pKa = 3.36QGEE57 pKa = 4.72VVLSTFPPGAAVLGSATTDD76 pKa = 2.95TSGRR80 pKa = 11.84FSIQTTPIDD89 pKa = 3.66SAALYY94 pKa = 10.4QLTLTGGSSIDD105 pKa = 3.59FASGYY110 pKa = 10.26SISLSSLDD118 pKa = 3.66SLSTIVTGKK127 pKa = 8.12TLLSSPVTITIMTTLMANQAQGMINRR153 pKa = 11.84GAAPLASFQNALGLWSGLLGFDD175 pKa = 3.71PTTTPVTDD183 pKa = 3.77PTAGPEE189 pKa = 4.28SMGYY193 pKa = 8.07PAIYY197 pKa = 10.45GLILAGFSQMAMAIGVNDD215 pKa = 4.1HH216 pKa = 7.0LSPGTSNTLGLLSVLNSDD234 pKa = 3.83ISDD237 pKa = 3.72GVLDD241 pKa = 4.04GLKK244 pKa = 10.59PGDD247 pKa = 4.41PNPLTYY253 pKa = 9.53YY254 pKa = 10.29TYY256 pKa = 10.99TLTADD261 pKa = 3.83TLRR264 pKa = 11.84KK265 pKa = 10.19DD266 pKa = 3.41IADD269 pKa = 3.64STLEE273 pKa = 3.94FLWGDD278 pKa = 3.81RR279 pKa = 11.84NQSGLAPIDD288 pKa = 3.37VSGSVNALALDD299 pKa = 3.92SSSLFPGNPLPTLPDD314 pKa = 4.31PGSPLITVSSPVNNAYY330 pKa = 10.77YY331 pKa = 11.01NNTLPLVATASDD343 pKa = 3.5ALGIGLFSVTSTTLPLPVNPPGSTSIQTGVDD374 pKa = 2.76ILGIPDD380 pKa = 4.02GPLTTVFSAEE390 pKa = 3.94NLAGTVSTQTISLSIDD406 pKa = 3.02NTAPTLQNLSPISATSYY423 pKa = 10.29DD424 pKa = 3.18ICGSTNYY431 pKa = 9.87SVQVTGQMVDD441 pKa = 3.19TGSGPSRR448 pKa = 11.84IQVQEE453 pKa = 3.98TAPVSQAISAYY464 pKa = 7.81STVVNRR470 pKa = 11.84TTSNVSFYY478 pKa = 11.53VFAPVNSPCKK488 pKa = 10.28AVTFTYY494 pKa = 10.79NITVYY499 pKa = 10.71DD500 pKa = 3.78ALNNQRR506 pKa = 11.84TIPYY510 pKa = 10.74SMIIANN516 pKa = 3.85
Molecular weight: 53.05 kDa
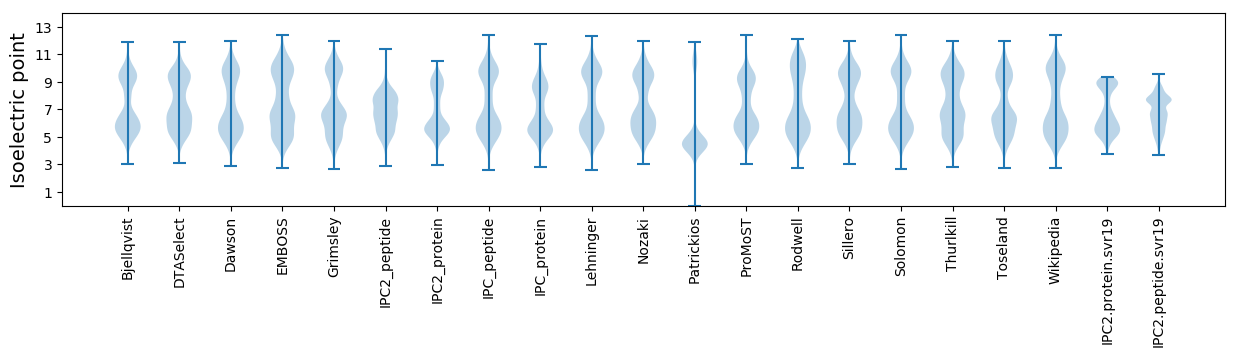
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I0IN68|I0IN68_LEPFC Putative cation transport protein OS=Leptospirillum ferrooxidans (strain C2-3) OX=1162668 GN=LFE_1014 PE=4 SV=1
MM1 pKa = 7.97WITIGALICFHH12 pKa = 6.84WGSHH16 pKa = 4.68LTSAFAVLGEE26 pKa = 4.47PEE28 pKa = 4.28SSVTADD34 pKa = 3.58RR35 pKa = 11.84EE36 pKa = 4.19RR37 pKa = 11.84SQGSHH42 pKa = 5.51SVRR45 pKa = 11.84TLPGGIRR52 pKa = 11.84MHH54 pKa = 7.28KK55 pKa = 8.08ITTAGGHH62 pKa = 4.65YY63 pKa = 10.08RR64 pKa = 11.84EE65 pKa = 5.37FSGQDD70 pKa = 3.21GAIFAITWRR79 pKa = 11.84GTKK82 pKa = 9.59PSHH85 pKa = 6.94LGDD88 pKa = 3.49LLGAYY93 pKa = 9.38RR94 pKa = 11.84QEE96 pKa = 4.61FIQAVRR102 pKa = 11.84TFRR105 pKa = 11.84MGKK108 pKa = 9.37APNRR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84GRR116 pKa = 11.84ILLIQTDD123 pKa = 3.82HH124 pKa = 7.61LLITGTGHH132 pKa = 6.89PRR134 pKa = 11.84DD135 pKa = 3.68LRR137 pKa = 11.84GMVEE141 pKa = 4.85IIGKK145 pKa = 6.68TPPGFDD151 pKa = 3.85PEE153 pKa = 4.91RR154 pKa = 11.84IPPQTPP160 pKa = 2.68
MM1 pKa = 7.97WITIGALICFHH12 pKa = 6.84WGSHH16 pKa = 4.68LTSAFAVLGEE26 pKa = 4.47PEE28 pKa = 4.28SSVTADD34 pKa = 3.58RR35 pKa = 11.84EE36 pKa = 4.19RR37 pKa = 11.84SQGSHH42 pKa = 5.51SVRR45 pKa = 11.84TLPGGIRR52 pKa = 11.84MHH54 pKa = 7.28KK55 pKa = 8.08ITTAGGHH62 pKa = 4.65YY63 pKa = 10.08RR64 pKa = 11.84EE65 pKa = 5.37FSGQDD70 pKa = 3.21GAIFAITWRR79 pKa = 11.84GTKK82 pKa = 9.59PSHH85 pKa = 6.94LGDD88 pKa = 3.49LLGAYY93 pKa = 9.38RR94 pKa = 11.84QEE96 pKa = 4.61FIQAVRR102 pKa = 11.84TFRR105 pKa = 11.84MGKK108 pKa = 9.37APNRR112 pKa = 11.84RR113 pKa = 11.84RR114 pKa = 11.84GRR116 pKa = 11.84ILLIQTDD123 pKa = 3.82HH124 pKa = 7.61LLITGTGHH132 pKa = 6.89PRR134 pKa = 11.84DD135 pKa = 3.68LRR137 pKa = 11.84GMVEE141 pKa = 4.85IIGKK145 pKa = 6.68TPPGFDD151 pKa = 3.85PEE153 pKa = 4.91RR154 pKa = 11.84IPPQTPP160 pKa = 2.68
Molecular weight: 17.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
742622 |
40 |
1727 |
307.8 |
34.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.122 ± 0.04 | 0.791 ± 0.017 |
5.024 ± 0.032 | 6.412 ± 0.055 |
4.458 ± 0.037 | 8.074 ± 0.052 |
2.295 ± 0.022 | 6.487 ± 0.04 |
5.003 ± 0.038 | 10.877 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.669 ± 0.022 | 3.156 ± 0.031 |
5.307 ± 0.034 | 3.105 ± 0.031 |
6.097 ± 0.044 | 7.76 ± 0.049 |
4.951 ± 0.038 | 6.763 ± 0.049 |
1.219 ± 0.02 | 2.429 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
