
Virus sp. ctgoX21
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.3
Get precalculated fractions of proteins
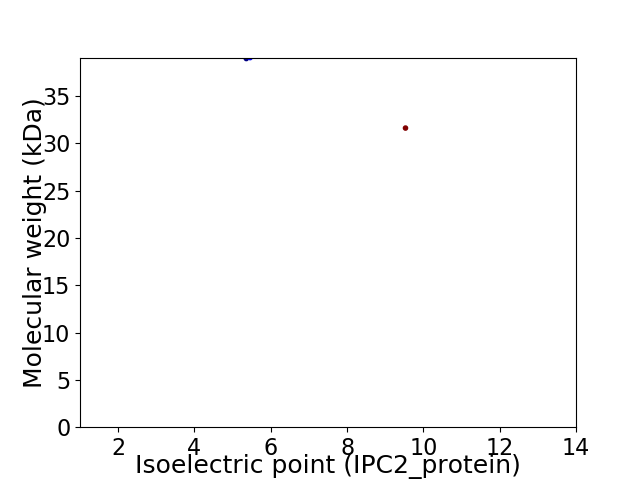
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2WBD0|A0A5Q2WBD0_9VIRU Rep protein OS=Virus sp. ctgoX21 OX=2656731 PE=3 SV=1
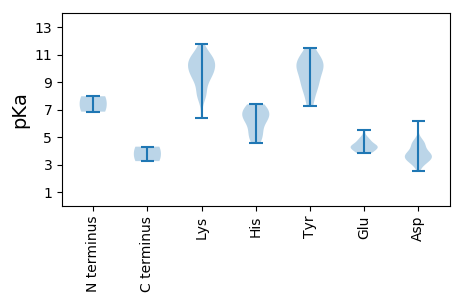
MM1 pKa = 7.97SDD3 pKa = 4.47LLTNLPTTVDD13 pKa = 3.51DD14 pKa = 4.5KK15 pKa = 11.57FRR17 pKa = 11.84LQAQRR22 pKa = 11.84LMLTYY27 pKa = 8.86KK28 pKa = 9.19THH30 pKa = 6.74LDD32 pKa = 3.21KK33 pKa = 11.78DD34 pKa = 3.91EE35 pKa = 4.74FSEE38 pKa = 4.18FLSRR42 pKa = 11.84IKK44 pKa = 10.33PPKK47 pKa = 7.99TLYY50 pKa = 9.71IAHH53 pKa = 7.08EE54 pKa = 4.7NGDD57 pKa = 4.45DD58 pKa = 4.81DD59 pKa = 6.17PITPYY64 pKa = 10.8EE65 pKa = 4.17HH66 pKa = 5.43THH68 pKa = 4.58VVIDD72 pKa = 4.56FGTAFNNRR80 pKa = 11.84NCRR83 pKa = 11.84VFDD86 pKa = 3.85FEE88 pKa = 5.54GIHH91 pKa = 5.27PHH93 pKa = 6.36ISKK96 pKa = 9.78IQNKK100 pKa = 9.24FDD102 pKa = 3.32WKK104 pKa = 10.24RR105 pKa = 11.84ACTYY109 pKa = 7.37VTKK112 pKa = 10.66EE113 pKa = 3.91DD114 pKa = 3.81TTVEE118 pKa = 3.9LAEE121 pKa = 4.35EE122 pKa = 4.4DD123 pKa = 3.76SFGVKK128 pKa = 9.94KK129 pKa = 10.87NSVLDD134 pKa = 3.14IWRR137 pKa = 11.84SRR139 pKa = 11.84NISEE143 pKa = 4.36ALEE146 pKa = 4.44GMSSLRR152 pKa = 11.84DD153 pKa = 3.82ALPTIAIFNHH163 pKa = 6.24KK164 pKa = 9.71PIEE167 pKa = 4.27LPQPEE172 pKa = 4.69LEE174 pKa = 4.99EE175 pKa = 4.2SQFWYY180 pKa = 8.52WQEE183 pKa = 3.8SCRR186 pKa = 11.84DD187 pKa = 3.89LIFTQPNNRR196 pKa = 11.84SVIWVYY202 pKa = 11.5DD203 pKa = 3.76EE204 pKa = 4.21VGGAGKK210 pKa = 8.21TRR212 pKa = 11.84FAEE215 pKa = 4.76WACLTYY221 pKa = 10.25PEE223 pKa = 4.97KK224 pKa = 10.68CTMFNQIGKK233 pKa = 9.97VSDD236 pKa = 3.36FAMNIRR242 pKa = 11.84SEE244 pKa = 4.12QLAGWRR250 pKa = 11.84GDD252 pKa = 3.33TCFINLSRR260 pKa = 11.84DD261 pKa = 3.63YY262 pKa = 11.14SDD264 pKa = 4.64KK265 pKa = 11.29DD266 pKa = 3.45HH267 pKa = 7.37IYY269 pKa = 10.4QALEE273 pKa = 3.89MIKK276 pKa = 10.57DD277 pKa = 3.73RR278 pKa = 11.84FVTCTKK284 pKa = 9.65YY285 pKa = 9.46TGGKK289 pKa = 7.99IWLPTMHH296 pKa = 6.88VIVLANFPPMKK307 pKa = 10.25HH308 pKa = 6.39KK309 pKa = 10.56LSKK312 pKa = 10.64DD313 pKa = 2.5RR314 pKa = 11.84WEE316 pKa = 4.45IYY318 pKa = 10.41EE319 pKa = 4.25IDD321 pKa = 3.56PTEE324 pKa = 3.95WSWIEE329 pKa = 3.86RR330 pKa = 11.84SSGG333 pKa = 3.25
MM1 pKa = 7.97SDD3 pKa = 4.47LLTNLPTTVDD13 pKa = 3.51DD14 pKa = 4.5KK15 pKa = 11.57FRR17 pKa = 11.84LQAQRR22 pKa = 11.84LMLTYY27 pKa = 8.86KK28 pKa = 9.19THH30 pKa = 6.74LDD32 pKa = 3.21KK33 pKa = 11.78DD34 pKa = 3.91EE35 pKa = 4.74FSEE38 pKa = 4.18FLSRR42 pKa = 11.84IKK44 pKa = 10.33PPKK47 pKa = 7.99TLYY50 pKa = 9.71IAHH53 pKa = 7.08EE54 pKa = 4.7NGDD57 pKa = 4.45DD58 pKa = 4.81DD59 pKa = 6.17PITPYY64 pKa = 10.8EE65 pKa = 4.17HH66 pKa = 5.43THH68 pKa = 4.58VVIDD72 pKa = 4.56FGTAFNNRR80 pKa = 11.84NCRR83 pKa = 11.84VFDD86 pKa = 3.85FEE88 pKa = 5.54GIHH91 pKa = 5.27PHH93 pKa = 6.36ISKK96 pKa = 9.78IQNKK100 pKa = 9.24FDD102 pKa = 3.32WKK104 pKa = 10.24RR105 pKa = 11.84ACTYY109 pKa = 7.37VTKK112 pKa = 10.66EE113 pKa = 3.91DD114 pKa = 3.81TTVEE118 pKa = 3.9LAEE121 pKa = 4.35EE122 pKa = 4.4DD123 pKa = 3.76SFGVKK128 pKa = 9.94KK129 pKa = 10.87NSVLDD134 pKa = 3.14IWRR137 pKa = 11.84SRR139 pKa = 11.84NISEE143 pKa = 4.36ALEE146 pKa = 4.44GMSSLRR152 pKa = 11.84DD153 pKa = 3.82ALPTIAIFNHH163 pKa = 6.24KK164 pKa = 9.71PIEE167 pKa = 4.27LPQPEE172 pKa = 4.69LEE174 pKa = 4.99EE175 pKa = 4.2SQFWYY180 pKa = 8.52WQEE183 pKa = 3.8SCRR186 pKa = 11.84DD187 pKa = 3.89LIFTQPNNRR196 pKa = 11.84SVIWVYY202 pKa = 11.5DD203 pKa = 3.76EE204 pKa = 4.21VGGAGKK210 pKa = 8.21TRR212 pKa = 11.84FAEE215 pKa = 4.76WACLTYY221 pKa = 10.25PEE223 pKa = 4.97KK224 pKa = 10.68CTMFNQIGKK233 pKa = 9.97VSDD236 pKa = 3.36FAMNIRR242 pKa = 11.84SEE244 pKa = 4.12QLAGWRR250 pKa = 11.84GDD252 pKa = 3.33TCFINLSRR260 pKa = 11.84DD261 pKa = 3.63YY262 pKa = 11.14SDD264 pKa = 4.64KK265 pKa = 11.29DD266 pKa = 3.45HH267 pKa = 7.37IYY269 pKa = 10.4QALEE273 pKa = 3.89MIKK276 pKa = 10.57DD277 pKa = 3.73RR278 pKa = 11.84FVTCTKK284 pKa = 9.65YY285 pKa = 9.46TGGKK289 pKa = 7.99IWLPTMHH296 pKa = 6.88VIVLANFPPMKK307 pKa = 10.25HH308 pKa = 6.39KK309 pKa = 10.56LSKK312 pKa = 10.64DD313 pKa = 2.5RR314 pKa = 11.84WEE316 pKa = 4.45IYY318 pKa = 10.41EE319 pKa = 4.25IDD321 pKa = 3.56PTEE324 pKa = 3.95WSWIEE329 pKa = 3.86RR330 pKa = 11.84SSGG333 pKa = 3.25
Molecular weight: 38.98 kDa
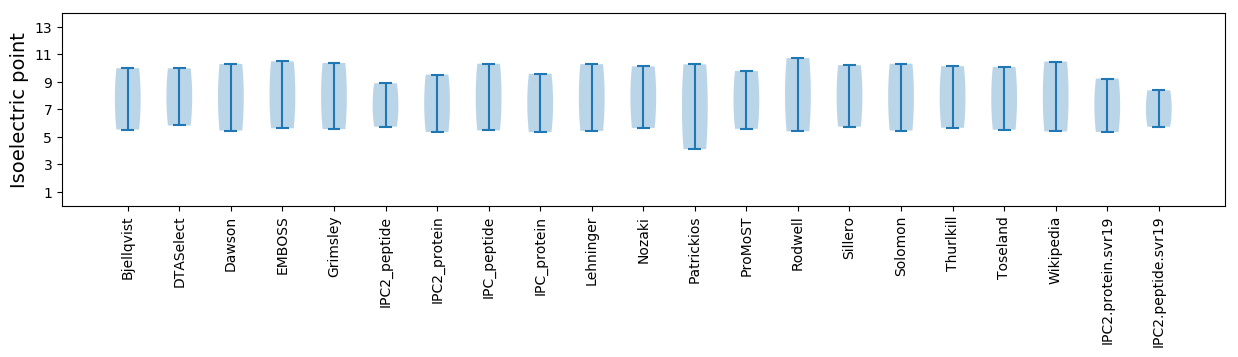
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2WBD0|A0A5Q2WBD0_9VIRU Rep protein OS=Virus sp. ctgoX21 OX=2656731 PE=3 SV=1
MM1 pKa = 6.85YY2 pKa = 9.11PRR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.97KK7 pKa = 8.98YY8 pKa = 8.15TPKK11 pKa = 9.87PFVRR15 pKa = 11.84RR16 pKa = 11.84SQKK19 pKa = 9.9PRR21 pKa = 11.84TAPRR25 pKa = 11.84RR26 pKa = 11.84KK27 pKa = 8.08YY28 pKa = 8.92TSKK31 pKa = 10.9KK32 pKa = 7.87VIKK35 pKa = 9.6RR36 pKa = 11.84VVRR39 pKa = 11.84AVVSRR44 pKa = 11.84QAEE47 pKa = 4.43KK48 pKa = 10.82KK49 pKa = 9.04SVVYY53 pKa = 10.69SSTLSLGTLQPTTSSLVGNYY73 pKa = 7.28MVITPSQSSFGYY85 pKa = 9.0TIAKK89 pKa = 8.2GTDD92 pKa = 2.93DD93 pKa = 3.61NQRR96 pKa = 11.84IGNQINIKK104 pKa = 6.41TLRR107 pKa = 11.84HH108 pKa = 4.98TFVCIVNPYY117 pKa = 10.03NATTNTQQRR126 pKa = 11.84PCFIRR131 pKa = 11.84FYY133 pKa = 9.89YY134 pKa = 10.27FKK136 pKa = 10.99SKK138 pKa = 9.62WSPVADD144 pKa = 3.3VATGNVCGVNANFFDD159 pKa = 4.16NSSSDD164 pKa = 3.35TGFSGALMDD173 pKa = 4.37LTRR176 pKa = 11.84KK177 pKa = 9.14IQSQNYY183 pKa = 8.28TYY185 pKa = 10.91LCHH188 pKa = 6.34RR189 pKa = 11.84TFKK192 pKa = 10.45IGNSQPAAGMATSSIQANQTSNDD215 pKa = 3.46FKK217 pKa = 10.64MSMIGKK223 pKa = 8.94VNLARR228 pKa = 11.84FCKK231 pKa = 10.59SKK233 pKa = 9.72FTFNDD238 pKa = 2.97ANQVMNPWIHH248 pKa = 7.16CLMQVMAADD257 pKa = 4.55GSVISTTQSQVSIQSQIYY275 pKa = 9.93CEE277 pKa = 4.26YY278 pKa = 10.21TDD280 pKa = 3.6EE281 pKa = 4.3
MM1 pKa = 6.85YY2 pKa = 9.11PRR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.97KK7 pKa = 8.98YY8 pKa = 8.15TPKK11 pKa = 9.87PFVRR15 pKa = 11.84RR16 pKa = 11.84SQKK19 pKa = 9.9PRR21 pKa = 11.84TAPRR25 pKa = 11.84RR26 pKa = 11.84KK27 pKa = 8.08YY28 pKa = 8.92TSKK31 pKa = 10.9KK32 pKa = 7.87VIKK35 pKa = 9.6RR36 pKa = 11.84VVRR39 pKa = 11.84AVVSRR44 pKa = 11.84QAEE47 pKa = 4.43KK48 pKa = 10.82KK49 pKa = 9.04SVVYY53 pKa = 10.69SSTLSLGTLQPTTSSLVGNYY73 pKa = 7.28MVITPSQSSFGYY85 pKa = 9.0TIAKK89 pKa = 8.2GTDD92 pKa = 2.93DD93 pKa = 3.61NQRR96 pKa = 11.84IGNQINIKK104 pKa = 6.41TLRR107 pKa = 11.84HH108 pKa = 4.98TFVCIVNPYY117 pKa = 10.03NATTNTQQRR126 pKa = 11.84PCFIRR131 pKa = 11.84FYY133 pKa = 9.89YY134 pKa = 10.27FKK136 pKa = 10.99SKK138 pKa = 9.62WSPVADD144 pKa = 3.3VATGNVCGVNANFFDD159 pKa = 4.16NSSSDD164 pKa = 3.35TGFSGALMDD173 pKa = 4.37LTRR176 pKa = 11.84KK177 pKa = 9.14IQSQNYY183 pKa = 8.28TYY185 pKa = 10.91LCHH188 pKa = 6.34RR189 pKa = 11.84TFKK192 pKa = 10.45IGNSQPAAGMATSSIQANQTSNDD215 pKa = 3.46FKK217 pKa = 10.64MSMIGKK223 pKa = 8.94VNLARR228 pKa = 11.84FCKK231 pKa = 10.59SKK233 pKa = 9.72FTFNDD238 pKa = 2.97ANQVMNPWIHH248 pKa = 7.16CLMQVMAADD257 pKa = 4.55GSVISTTQSQVSIQSQIYY275 pKa = 9.93CEE277 pKa = 4.26YY278 pKa = 10.21TDD280 pKa = 3.6EE281 pKa = 4.3
Molecular weight: 31.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
614 |
281 |
333 |
307.0 |
35.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.212 ± 0.543 | 2.28 ± 0.137 |
5.7 ± 1.387 | 4.56 ± 2.262 |
5.212 ± 0.149 | 4.397 ± 0.148 |
2.117 ± 0.68 | 6.515 ± 0.532 |
6.84 ± 0.179 | 5.537 ± 1.282 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.769 ± 0.281 | 5.537 ± 0.793 |
4.56 ± 0.188 | 4.723 ± 1.32 |
5.7 ± 0.226 | 8.306 ± 1.535 |
8.143 ± 0.718 | 5.863 ± 1.043 |
2.117 ± 0.91 | 3.909 ± 0.465 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
