
Odontoglossum ringspot virus (isolate Singapore 1) (ORSV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Tobamovirus; Odontoglossum ringspot virus
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
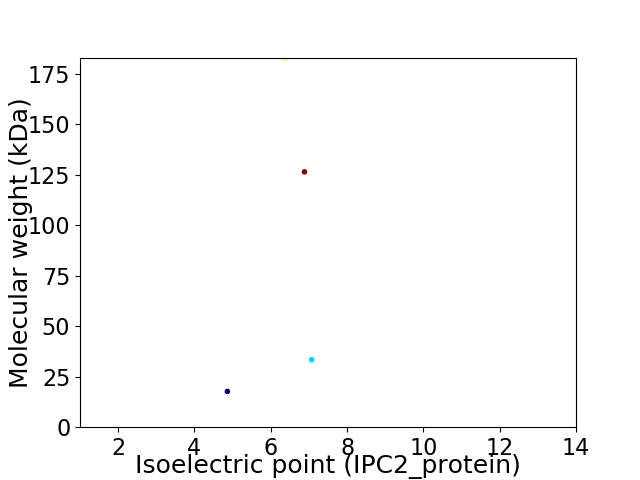
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
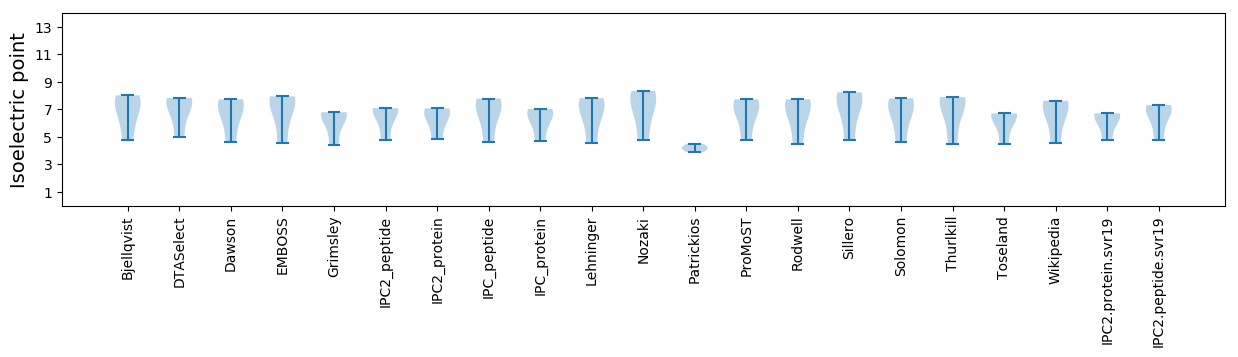
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q84136|CAPSD_ORSVS Capsid protein OS=Odontoglossum ringspot virus (isolate Singapore 1) OX=138662 GN=CP PE=3 SV=3
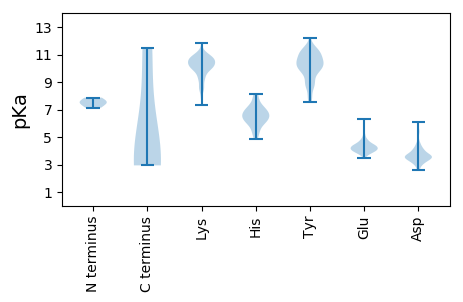
MM1 pKa = 7.87SYY3 pKa = 10.61TITDD7 pKa = 3.91PSKK10 pKa = 10.62LAYY13 pKa = 10.2LSSAWADD20 pKa = 3.42PNSLINLCTNSLGNQFQTQQARR42 pKa = 11.84TTVQQQFADD51 pKa = 3.29VWQPVPTLTSRR62 pKa = 11.84FPAGAGYY69 pKa = 9.93FRR71 pKa = 11.84VYY73 pKa = 10.28RR74 pKa = 11.84YY75 pKa = 10.74DD76 pKa = 5.0PILDD80 pKa = 3.81PLITFLMGTFDD91 pKa = 3.16TRR93 pKa = 11.84NRR95 pKa = 11.84IIEE98 pKa = 4.12VEE100 pKa = 4.11NPQNPTTTEE109 pKa = 3.82TLDD112 pKa = 2.98ATRR115 pKa = 11.84RR116 pKa = 11.84VDD118 pKa = 3.5DD119 pKa = 3.56ATVAIRR125 pKa = 11.84SAINNLLNEE134 pKa = 4.65LVRR137 pKa = 11.84GTGMYY142 pKa = 10.06NQVSFEE148 pKa = 4.28TMSGLTWTSSS158 pKa = 2.94
MM1 pKa = 7.87SYY3 pKa = 10.61TITDD7 pKa = 3.91PSKK10 pKa = 10.62LAYY13 pKa = 10.2LSSAWADD20 pKa = 3.42PNSLINLCTNSLGNQFQTQQARR42 pKa = 11.84TTVQQQFADD51 pKa = 3.29VWQPVPTLTSRR62 pKa = 11.84FPAGAGYY69 pKa = 9.93FRR71 pKa = 11.84VYY73 pKa = 10.28RR74 pKa = 11.84YY75 pKa = 10.74DD76 pKa = 5.0PILDD80 pKa = 3.81PLITFLMGTFDD91 pKa = 3.16TRR93 pKa = 11.84NRR95 pKa = 11.84IIEE98 pKa = 4.12VEE100 pKa = 4.11NPQNPTTTEE109 pKa = 3.82TLDD112 pKa = 2.98ATRR115 pKa = 11.84RR116 pKa = 11.84VDD118 pKa = 3.5DD119 pKa = 3.56ATVAIRR125 pKa = 11.84SAINNLLNEE134 pKa = 4.65LVRR137 pKa = 11.84GTGMYY142 pKa = 10.06NQVSFEE148 pKa = 4.28TMSGLTWTSSS158 pKa = 2.94
Molecular weight: 17.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q84136|CAPSD_ORSVS Capsid protein OS=Odontoglossum ringspot virus (isolate Singapore 1) OX=138662 GN=CP PE=3 SV=3
MM1 pKa = 7.13GRR3 pKa = 11.84LRR5 pKa = 11.84FVVLLSIFPIKK16 pKa = 9.79TFSEE20 pKa = 4.67PCSTMALVLRR30 pKa = 11.84DD31 pKa = 3.82SIKK34 pKa = 10.14ISEE37 pKa = 5.09FINLSASEE45 pKa = 4.28KK46 pKa = 10.15LLPSALTAVKK56 pKa = 9.87SVRR59 pKa = 11.84ISKK62 pKa = 9.58VDD64 pKa = 3.69KK65 pKa = 10.57IISYY69 pKa = 10.88EE70 pKa = 3.98NDD72 pKa = 3.31TLSDD76 pKa = 3.3IDD78 pKa = 3.91LLKK81 pKa = 10.73GVKK84 pKa = 9.67LVEE87 pKa = 3.99NGYY90 pKa = 9.81VCLAGLVVTGEE101 pKa = 3.84WNLPDD106 pKa = 3.51NCKK109 pKa = 10.69GGVSICLVDD118 pKa = 4.54KK119 pKa = 10.87RR120 pKa = 11.84MKK122 pKa = 10.14RR123 pKa = 11.84ANEE126 pKa = 3.84ATLGSYY132 pKa = 7.53HH133 pKa = 6.79TSACKK138 pKa = 10.09KK139 pKa = 9.82RR140 pKa = 11.84FTFKK144 pKa = 10.61IIPNYY149 pKa = 10.75SVTTADD155 pKa = 3.35ALKK158 pKa = 10.81GIWQVMINIRR168 pKa = 11.84GVEE171 pKa = 4.04MEE173 pKa = 5.2KK174 pKa = 10.84GFCPLSLEE182 pKa = 4.19FVSICVVYY190 pKa = 10.66LNNIKK195 pKa = 10.6LGLRR199 pKa = 11.84EE200 pKa = 4.4KK201 pKa = 10.37ILNVTEE207 pKa = 4.6GGPTEE212 pKa = 4.04LTEE215 pKa = 4.07AVVDD219 pKa = 3.87EE220 pKa = 4.88FVEE223 pKa = 4.36KK224 pKa = 11.04VPMAARR230 pKa = 11.84LKK232 pKa = 10.71SFRR235 pKa = 11.84SVNKK239 pKa = 9.88KK240 pKa = 10.34KK241 pKa = 10.45PSDD244 pKa = 3.34SSKK247 pKa = 10.51FVNGKK252 pKa = 9.57SRR254 pKa = 11.84LNSRR258 pKa = 11.84NKK260 pKa = 10.32LNYY263 pKa = 9.72EE264 pKa = 4.14NGDD267 pKa = 3.54SDD269 pKa = 4.01VGISVVDD276 pKa = 4.56DD277 pKa = 3.39IVVGNGVSDD286 pKa = 3.3IRR288 pKa = 11.84IDD290 pKa = 4.04DD291 pKa = 4.05DD292 pKa = 4.61CEE294 pKa = 4.88SFDD297 pKa = 4.49AQSDD301 pKa = 4.35PYY303 pKa = 11.45
MM1 pKa = 7.13GRR3 pKa = 11.84LRR5 pKa = 11.84FVVLLSIFPIKK16 pKa = 9.79TFSEE20 pKa = 4.67PCSTMALVLRR30 pKa = 11.84DD31 pKa = 3.82SIKK34 pKa = 10.14ISEE37 pKa = 5.09FINLSASEE45 pKa = 4.28KK46 pKa = 10.15LLPSALTAVKK56 pKa = 9.87SVRR59 pKa = 11.84ISKK62 pKa = 9.58VDD64 pKa = 3.69KK65 pKa = 10.57IISYY69 pKa = 10.88EE70 pKa = 3.98NDD72 pKa = 3.31TLSDD76 pKa = 3.3IDD78 pKa = 3.91LLKK81 pKa = 10.73GVKK84 pKa = 9.67LVEE87 pKa = 3.99NGYY90 pKa = 9.81VCLAGLVVTGEE101 pKa = 3.84WNLPDD106 pKa = 3.51NCKK109 pKa = 10.69GGVSICLVDD118 pKa = 4.54KK119 pKa = 10.87RR120 pKa = 11.84MKK122 pKa = 10.14RR123 pKa = 11.84ANEE126 pKa = 3.84ATLGSYY132 pKa = 7.53HH133 pKa = 6.79TSACKK138 pKa = 10.09KK139 pKa = 9.82RR140 pKa = 11.84FTFKK144 pKa = 10.61IIPNYY149 pKa = 10.75SVTTADD155 pKa = 3.35ALKK158 pKa = 10.81GIWQVMINIRR168 pKa = 11.84GVEE171 pKa = 4.04MEE173 pKa = 5.2KK174 pKa = 10.84GFCPLSLEE182 pKa = 4.19FVSICVVYY190 pKa = 10.66LNNIKK195 pKa = 10.6LGLRR199 pKa = 11.84EE200 pKa = 4.4KK201 pKa = 10.37ILNVTEE207 pKa = 4.6GGPTEE212 pKa = 4.04LTEE215 pKa = 4.07AVVDD219 pKa = 3.87EE220 pKa = 4.88FVEE223 pKa = 4.36KK224 pKa = 11.04VPMAARR230 pKa = 11.84LKK232 pKa = 10.71SFRR235 pKa = 11.84SVNKK239 pKa = 9.88KK240 pKa = 10.34KK241 pKa = 10.45PSDD244 pKa = 3.34SSKK247 pKa = 10.51FVNGKK252 pKa = 9.57SRR254 pKa = 11.84LNSRR258 pKa = 11.84NKK260 pKa = 10.32LNYY263 pKa = 9.72EE264 pKa = 4.14NGDD267 pKa = 3.54SDD269 pKa = 4.01VGISVVDD276 pKa = 4.56DD277 pKa = 3.39IVVGNGVSDD286 pKa = 3.3IRR288 pKa = 11.84IDD290 pKa = 4.04DD291 pKa = 4.05DD292 pKa = 4.61CEE294 pKa = 4.88SFDD297 pKa = 4.49AQSDD301 pKa = 4.35PYY303 pKa = 11.45
Molecular weight: 33.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3185 |
158 |
1612 |
796.3 |
90.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.934 ± 0.255 | 2.135 ± 0.202 |
6.499 ± 0.107 | 5.746 ± 0.329 |
5.243 ± 0.252 | 4.898 ± 0.253 |
1.947 ± 0.384 | 5.275 ± 0.405 |
7.567 ± 0.877 | 9.294 ± 0.089 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.512 ± 0.108 | 4.584 ± 0.396 |
4.05 ± 0.242 | 3.234 ± 0.573 |
4.584 ± 0.244 | 7.63 ± 0.39 |
5.651 ± 0.974 | 8.289 ± 0.592 |
0.973 ± 0.126 | 3.925 ± 0.282 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
