
Heterobasidion partitivirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Alphapartitivirus
Average proteome isoelectric point is 6.79
Get precalculated fractions of proteins
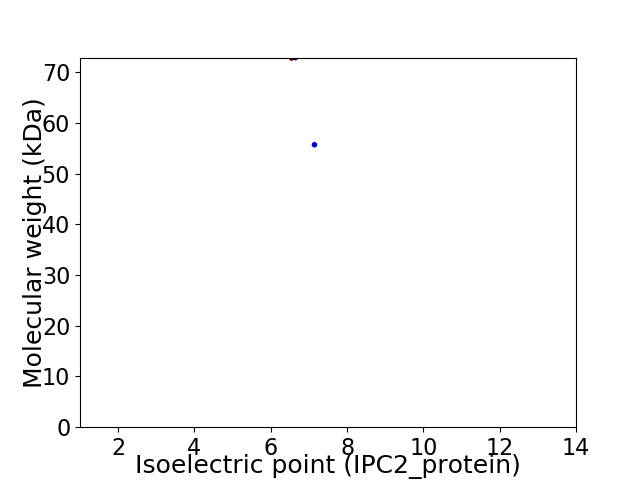
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E7E1E9|E7E1E9_9VIRU Putative coat protein OS=Heterobasidion partitivirus 1 OX=942041 PE=4 SV=1
MM1 pKa = 8.34DD2 pKa = 4.58YY3 pKa = 10.36LTGLFSRR10 pKa = 11.84VLHH13 pKa = 6.7ISRR16 pKa = 11.84KK17 pKa = 5.46VTNFEE22 pKa = 3.98FAGTYY27 pKa = 9.64HH28 pKa = 5.98YY29 pKa = 10.51QPSIPQVNEE38 pKa = 3.7VATEE42 pKa = 3.8NHH44 pKa = 6.09KK45 pKa = 9.9RR46 pKa = 11.84TLRR49 pKa = 11.84HH50 pKa = 5.27SFRR53 pKa = 11.84TYY55 pKa = 9.43LTSDD59 pKa = 3.65EE60 pKa = 4.02YY61 pKa = 11.8DD62 pKa = 3.88KK63 pKa = 11.21IVNGYY68 pKa = 9.7KK69 pKa = 8.98RR70 pKa = 11.84TNLDD74 pKa = 3.15PSTITEE80 pKa = 4.51DD81 pKa = 3.62FFSGDD86 pKa = 3.46IEE88 pKa = 4.39DD89 pKa = 4.52HH90 pKa = 7.09PEE92 pKa = 3.86PTDD95 pKa = 3.65FKK97 pKa = 10.44SQLSIEE103 pKa = 4.45YY104 pKa = 10.13GLQCMIDD111 pKa = 3.7AFKK114 pKa = 10.53PPAPARR120 pKa = 11.84VCHH123 pKa = 6.74LYY125 pKa = 10.49DD126 pKa = 3.94VQWHH130 pKa = 5.98YY131 pKa = 10.81PFKK134 pKa = 9.3WQVNAEE140 pKa = 4.24APFSTEE146 pKa = 4.86KK147 pKa = 11.1YY148 pKa = 10.6FLDD151 pKa = 3.51LRR153 pKa = 11.84KK154 pKa = 10.25KK155 pKa = 10.72FGDD158 pKa = 3.47FFDD161 pKa = 4.94PVTKK165 pKa = 10.53LWTKK169 pKa = 9.96YY170 pKa = 10.08VNPLDD175 pKa = 3.5ALRR178 pKa = 11.84RR179 pKa = 11.84YY180 pKa = 9.53GHH182 pKa = 6.4TPPADD187 pKa = 3.57TLNQVTPPKK196 pKa = 10.31FGFMKK201 pKa = 10.58NLIFSFVHH209 pKa = 5.54SWQHH213 pKa = 4.84VIKK216 pKa = 10.93SRR218 pKa = 11.84FTSNAGITHH227 pKa = 6.49SNFLRR232 pKa = 11.84QRR234 pKa = 11.84FLFPMLLHH242 pKa = 6.79IKK244 pKa = 8.41TAIVSFDD251 pKa = 3.68APNKK255 pKa = 9.89LRR257 pKa = 11.84SIWGVSKK264 pKa = 10.63LWIISEE270 pKa = 3.97AMIYY274 pKa = 9.27WEE276 pKa = 4.13YY277 pKa = 10.51IAWIKK282 pKa = 10.29LNPGSTPMLWGYY294 pKa = 7.52EE295 pKa = 4.17TFTGGWFRR303 pKa = 11.84LWRR306 pKa = 11.84DD307 pKa = 3.02LHH309 pKa = 6.21TPGEE313 pKa = 4.43DD314 pKa = 2.98VTYY317 pKa = 9.36ITIDD321 pKa = 2.75WSRR324 pKa = 11.84FDD326 pKa = 3.08KK327 pKa = 10.82RR328 pKa = 11.84AYY330 pKa = 9.23FWLIRR335 pKa = 11.84KK336 pKa = 9.19IFIRR340 pKa = 11.84TRR342 pKa = 11.84CFLDD346 pKa = 3.42FTNGYY351 pKa = 10.01VSTKK355 pKa = 10.21DD356 pKa = 3.7YY357 pKa = 7.47PTSPTDD363 pKa = 3.58PDD365 pKa = 3.34KK366 pKa = 11.52LQALWEE372 pKa = 4.09WTIEE376 pKa = 4.06AFFDD380 pKa = 4.15SPIVLPDD387 pKa = 2.96GSMFKK392 pKa = 10.54RR393 pKa = 11.84LFAGIPSGLFITQLMDD409 pKa = 2.64SWYY412 pKa = 10.28NYY414 pKa = 9.21TMLAAILHH422 pKa = 5.01YY423 pKa = 9.74MGYY426 pKa = 10.49DD427 pKa = 3.37PRR429 pKa = 11.84RR430 pKa = 11.84CIIKK434 pKa = 10.07VQGDD438 pKa = 3.48DD439 pKa = 3.95SIIRR443 pKa = 11.84LYY445 pKa = 10.31IQIPLHH451 pKa = 5.24EE452 pKa = 5.68HH453 pKa = 6.91DD454 pKa = 5.42LFLLRR459 pKa = 11.84MQEE462 pKa = 4.1VSDD465 pKa = 3.89HH466 pKa = 6.52LFGAKK471 pKa = 9.25ISFEE475 pKa = 3.94KK476 pKa = 10.94SEE478 pKa = 4.55LRR480 pKa = 11.84NSLIGSEE487 pKa = 4.05VLSYY491 pKa = 11.31RR492 pKa = 11.84NIQGLPYY499 pKa = 9.77RR500 pKa = 11.84DD501 pKa = 4.35LIKK504 pKa = 10.32MLAQFYY510 pKa = 7.91HH511 pKa = 6.55TKK513 pKa = 10.76AKK515 pKa = 10.74DD516 pKa = 3.4PTPEE520 pKa = 3.31ITMAQAIGFAYY531 pKa = 9.62AACGNDD537 pKa = 3.41FRR539 pKa = 11.84IHH541 pKa = 5.78EE542 pKa = 4.71LLRR545 pKa = 11.84SVYY548 pKa = 10.21DD549 pKa = 3.44YY550 pKa = 11.77YY551 pKa = 11.07KK552 pKa = 11.03AQGFTPNPAGLTVVFGDD569 pKa = 3.79SPDD572 pKa = 3.7RR573 pKa = 11.84PDD575 pKa = 3.59YY576 pKa = 10.69PISLDD581 pKa = 3.65EE582 pKa = 5.37FPSQMDD588 pKa = 3.2VQRR591 pKa = 11.84FFLSTDD597 pKa = 3.05YY598 pKa = 11.41RR599 pKa = 11.84NADD602 pKa = 3.27QEE604 pKa = 4.11NRR606 pKa = 11.84TWPSSHH612 pKa = 7.01FLYY615 pKa = 10.62APCSRR620 pKa = 11.84II621 pKa = 3.44
MM1 pKa = 8.34DD2 pKa = 4.58YY3 pKa = 10.36LTGLFSRR10 pKa = 11.84VLHH13 pKa = 6.7ISRR16 pKa = 11.84KK17 pKa = 5.46VTNFEE22 pKa = 3.98FAGTYY27 pKa = 9.64HH28 pKa = 5.98YY29 pKa = 10.51QPSIPQVNEE38 pKa = 3.7VATEE42 pKa = 3.8NHH44 pKa = 6.09KK45 pKa = 9.9RR46 pKa = 11.84TLRR49 pKa = 11.84HH50 pKa = 5.27SFRR53 pKa = 11.84TYY55 pKa = 9.43LTSDD59 pKa = 3.65EE60 pKa = 4.02YY61 pKa = 11.8DD62 pKa = 3.88KK63 pKa = 11.21IVNGYY68 pKa = 9.7KK69 pKa = 8.98RR70 pKa = 11.84TNLDD74 pKa = 3.15PSTITEE80 pKa = 4.51DD81 pKa = 3.62FFSGDD86 pKa = 3.46IEE88 pKa = 4.39DD89 pKa = 4.52HH90 pKa = 7.09PEE92 pKa = 3.86PTDD95 pKa = 3.65FKK97 pKa = 10.44SQLSIEE103 pKa = 4.45YY104 pKa = 10.13GLQCMIDD111 pKa = 3.7AFKK114 pKa = 10.53PPAPARR120 pKa = 11.84VCHH123 pKa = 6.74LYY125 pKa = 10.49DD126 pKa = 3.94VQWHH130 pKa = 5.98YY131 pKa = 10.81PFKK134 pKa = 9.3WQVNAEE140 pKa = 4.24APFSTEE146 pKa = 4.86KK147 pKa = 11.1YY148 pKa = 10.6FLDD151 pKa = 3.51LRR153 pKa = 11.84KK154 pKa = 10.25KK155 pKa = 10.72FGDD158 pKa = 3.47FFDD161 pKa = 4.94PVTKK165 pKa = 10.53LWTKK169 pKa = 9.96YY170 pKa = 10.08VNPLDD175 pKa = 3.5ALRR178 pKa = 11.84RR179 pKa = 11.84YY180 pKa = 9.53GHH182 pKa = 6.4TPPADD187 pKa = 3.57TLNQVTPPKK196 pKa = 10.31FGFMKK201 pKa = 10.58NLIFSFVHH209 pKa = 5.54SWQHH213 pKa = 4.84VIKK216 pKa = 10.93SRR218 pKa = 11.84FTSNAGITHH227 pKa = 6.49SNFLRR232 pKa = 11.84QRR234 pKa = 11.84FLFPMLLHH242 pKa = 6.79IKK244 pKa = 8.41TAIVSFDD251 pKa = 3.68APNKK255 pKa = 9.89LRR257 pKa = 11.84SIWGVSKK264 pKa = 10.63LWIISEE270 pKa = 3.97AMIYY274 pKa = 9.27WEE276 pKa = 4.13YY277 pKa = 10.51IAWIKK282 pKa = 10.29LNPGSTPMLWGYY294 pKa = 7.52EE295 pKa = 4.17TFTGGWFRR303 pKa = 11.84LWRR306 pKa = 11.84DD307 pKa = 3.02LHH309 pKa = 6.21TPGEE313 pKa = 4.43DD314 pKa = 2.98VTYY317 pKa = 9.36ITIDD321 pKa = 2.75WSRR324 pKa = 11.84FDD326 pKa = 3.08KK327 pKa = 10.82RR328 pKa = 11.84AYY330 pKa = 9.23FWLIRR335 pKa = 11.84KK336 pKa = 9.19IFIRR340 pKa = 11.84TRR342 pKa = 11.84CFLDD346 pKa = 3.42FTNGYY351 pKa = 10.01VSTKK355 pKa = 10.21DD356 pKa = 3.7YY357 pKa = 7.47PTSPTDD363 pKa = 3.58PDD365 pKa = 3.34KK366 pKa = 11.52LQALWEE372 pKa = 4.09WTIEE376 pKa = 4.06AFFDD380 pKa = 4.15SPIVLPDD387 pKa = 2.96GSMFKK392 pKa = 10.54RR393 pKa = 11.84LFAGIPSGLFITQLMDD409 pKa = 2.64SWYY412 pKa = 10.28NYY414 pKa = 9.21TMLAAILHH422 pKa = 5.01YY423 pKa = 9.74MGYY426 pKa = 10.49DD427 pKa = 3.37PRR429 pKa = 11.84RR430 pKa = 11.84CIIKK434 pKa = 10.07VQGDD438 pKa = 3.48DD439 pKa = 3.95SIIRR443 pKa = 11.84LYY445 pKa = 10.31IQIPLHH451 pKa = 5.24EE452 pKa = 5.68HH453 pKa = 6.91DD454 pKa = 5.42LFLLRR459 pKa = 11.84MQEE462 pKa = 4.1VSDD465 pKa = 3.89HH466 pKa = 6.52LFGAKK471 pKa = 9.25ISFEE475 pKa = 3.94KK476 pKa = 10.94SEE478 pKa = 4.55LRR480 pKa = 11.84NSLIGSEE487 pKa = 4.05VLSYY491 pKa = 11.31RR492 pKa = 11.84NIQGLPYY499 pKa = 9.77RR500 pKa = 11.84DD501 pKa = 4.35LIKK504 pKa = 10.32MLAQFYY510 pKa = 7.91HH511 pKa = 6.55TKK513 pKa = 10.76AKK515 pKa = 10.74DD516 pKa = 3.4PTPEE520 pKa = 3.31ITMAQAIGFAYY531 pKa = 9.62AACGNDD537 pKa = 3.41FRR539 pKa = 11.84IHH541 pKa = 5.78EE542 pKa = 4.71LLRR545 pKa = 11.84SVYY548 pKa = 10.21DD549 pKa = 3.44YY550 pKa = 11.77YY551 pKa = 11.07KK552 pKa = 11.03AQGFTPNPAGLTVVFGDD569 pKa = 3.79SPDD572 pKa = 3.7RR573 pKa = 11.84PDD575 pKa = 3.59YY576 pKa = 10.69PISLDD581 pKa = 3.65EE582 pKa = 5.37FPSQMDD588 pKa = 3.2VQRR591 pKa = 11.84FFLSTDD597 pKa = 3.05YY598 pKa = 11.41RR599 pKa = 11.84NADD602 pKa = 3.27QEE604 pKa = 4.11NRR606 pKa = 11.84TWPSSHH612 pKa = 7.01FLYY615 pKa = 10.62APCSRR620 pKa = 11.84II621 pKa = 3.44
Molecular weight: 72.78 kDa
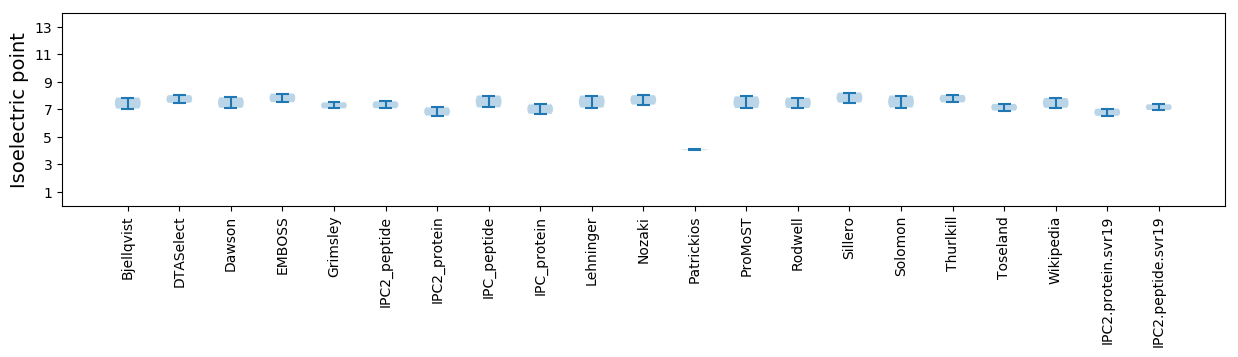
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E7E1E9|E7E1E9_9VIRU Putative coat protein OS=Heterobasidion partitivirus 1 OX=942041 PE=4 SV=1
MM1 pKa = 7.22AQNTVSAQVPTMEE14 pKa = 4.92PQPAGPGVAQPQVNLAPPVPDD35 pKa = 4.13PRR37 pKa = 11.84DD38 pKa = 3.45VAANAMMPQVPGPMPSSSGALSTITAALGYY68 pKa = 10.64NRR70 pKa = 11.84FLPMTFGRR78 pKa = 11.84STYY81 pKa = 8.95TPSFILLSLVLSEE94 pKa = 5.37FDD96 pKa = 3.64RR97 pKa = 11.84IMINTHH103 pKa = 6.14RR104 pKa = 11.84FYY106 pKa = 10.18QTSPEE111 pKa = 3.75WHH113 pKa = 7.24PIMSQLYY120 pKa = 8.19YY121 pKa = 10.23GVIFIVHH128 pKa = 6.43ILVARR133 pKa = 11.84RR134 pKa = 11.84TAQVISIDD142 pKa = 3.8EE143 pKa = 4.24NNFLDD148 pKa = 3.49WFEE151 pKa = 4.92TNFPLSSLPIAGPLKK166 pKa = 10.76HH167 pKa = 6.55FFQAITVTCGPSKK180 pKa = 10.82YY181 pKa = 9.97YY182 pKa = 11.19GNIYY186 pKa = 9.32PQLNTDD192 pKa = 3.0WTAAFANYY200 pKa = 9.83YY201 pKa = 9.54MPDD204 pKa = 3.18NAISNNHH211 pKa = 6.49LPPLPIFMDD220 pKa = 3.63MLNDD224 pKa = 4.26LLADD228 pKa = 4.0RR229 pKa = 11.84PGRR232 pKa = 11.84AATATVTPRR241 pKa = 11.84LAFDD245 pKa = 4.03HH246 pKa = 7.0RR247 pKa = 11.84NVDD250 pKa = 2.92HH251 pKa = 6.43WRR253 pKa = 11.84SYY255 pKa = 10.6YY256 pKa = 10.85LPLSHH261 pKa = 6.98TFSAMNNSSAVFQFAGMTSLHH282 pKa = 6.63ALPATAMPNWYY293 pKa = 9.31DD294 pKa = 2.99ASLFFGYY301 pKa = 9.71PNRR304 pKa = 11.84IDD306 pKa = 4.17GNALANISTIGEE318 pKa = 4.14FTRR321 pKa = 11.84LGTAGTQHH329 pKa = 6.19TLWFSRR335 pKa = 11.84TIQVMQRR342 pKa = 11.84HH343 pKa = 5.36AQFAKK348 pKa = 10.55DD349 pKa = 3.69STSLASISTKK359 pKa = 10.75GLGACLPILHH369 pKa = 7.02LSANAEE375 pKa = 3.94LSLPNATNNGQTVRR389 pKa = 11.84NVQAIAPNVTGAAAAAVATSPGRR412 pKa = 11.84PAGFAVARR420 pKa = 11.84PTHH423 pKa = 4.75FTVRR427 pKa = 11.84ATSKK431 pKa = 11.04LEE433 pKa = 3.96SLHH436 pKa = 7.02LLAEE440 pKa = 4.23QFSLLSCINLDD451 pKa = 3.65MTNLAARR458 pKa = 11.84GNAYY462 pKa = 10.08HH463 pKa = 7.47AFTTNAQVRR472 pKa = 11.84HH473 pKa = 5.55GPFWDD478 pKa = 3.65LADD481 pKa = 3.92VKK483 pKa = 11.05SHH485 pKa = 5.82TEE487 pKa = 3.75INVLGQISSFLAGAFHH503 pKa = 7.31VDD505 pKa = 3.25QRR507 pKa = 11.84QTKK510 pKa = 9.06
MM1 pKa = 7.22AQNTVSAQVPTMEE14 pKa = 4.92PQPAGPGVAQPQVNLAPPVPDD35 pKa = 4.13PRR37 pKa = 11.84DD38 pKa = 3.45VAANAMMPQVPGPMPSSSGALSTITAALGYY68 pKa = 10.64NRR70 pKa = 11.84FLPMTFGRR78 pKa = 11.84STYY81 pKa = 8.95TPSFILLSLVLSEE94 pKa = 5.37FDD96 pKa = 3.64RR97 pKa = 11.84IMINTHH103 pKa = 6.14RR104 pKa = 11.84FYY106 pKa = 10.18QTSPEE111 pKa = 3.75WHH113 pKa = 7.24PIMSQLYY120 pKa = 8.19YY121 pKa = 10.23GVIFIVHH128 pKa = 6.43ILVARR133 pKa = 11.84RR134 pKa = 11.84TAQVISIDD142 pKa = 3.8EE143 pKa = 4.24NNFLDD148 pKa = 3.49WFEE151 pKa = 4.92TNFPLSSLPIAGPLKK166 pKa = 10.76HH167 pKa = 6.55FFQAITVTCGPSKK180 pKa = 10.82YY181 pKa = 9.97YY182 pKa = 11.19GNIYY186 pKa = 9.32PQLNTDD192 pKa = 3.0WTAAFANYY200 pKa = 9.83YY201 pKa = 9.54MPDD204 pKa = 3.18NAISNNHH211 pKa = 6.49LPPLPIFMDD220 pKa = 3.63MLNDD224 pKa = 4.26LLADD228 pKa = 4.0RR229 pKa = 11.84PGRR232 pKa = 11.84AATATVTPRR241 pKa = 11.84LAFDD245 pKa = 4.03HH246 pKa = 7.0RR247 pKa = 11.84NVDD250 pKa = 2.92HH251 pKa = 6.43WRR253 pKa = 11.84SYY255 pKa = 10.6YY256 pKa = 10.85LPLSHH261 pKa = 6.98TFSAMNNSSAVFQFAGMTSLHH282 pKa = 6.63ALPATAMPNWYY293 pKa = 9.31DD294 pKa = 2.99ASLFFGYY301 pKa = 9.71PNRR304 pKa = 11.84IDD306 pKa = 4.17GNALANISTIGEE318 pKa = 4.14FTRR321 pKa = 11.84LGTAGTQHH329 pKa = 6.19TLWFSRR335 pKa = 11.84TIQVMQRR342 pKa = 11.84HH343 pKa = 5.36AQFAKK348 pKa = 10.55DD349 pKa = 3.69STSLASISTKK359 pKa = 10.75GLGACLPILHH369 pKa = 7.02LSANAEE375 pKa = 3.94LSLPNATNNGQTVRR389 pKa = 11.84NVQAIAPNVTGAAAAAVATSPGRR412 pKa = 11.84PAGFAVARR420 pKa = 11.84PTHH423 pKa = 4.75FTVRR427 pKa = 11.84ATSKK431 pKa = 11.04LEE433 pKa = 3.96SLHH436 pKa = 7.02LLAEE440 pKa = 4.23QFSLLSCINLDD451 pKa = 3.65MTNLAARR458 pKa = 11.84GNAYY462 pKa = 10.08HH463 pKa = 7.47AFTTNAQVRR472 pKa = 11.84HH473 pKa = 5.55GPFWDD478 pKa = 3.65LADD481 pKa = 3.92VKK483 pKa = 11.05SHH485 pKa = 5.82TEE487 pKa = 3.75INVLGQISSFLAGAFHH503 pKa = 7.31VDD505 pKa = 3.25QRR507 pKa = 11.84QTKK510 pKa = 9.06
Molecular weight: 55.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1131 |
510 |
621 |
565.5 |
64.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.311 ± 2.582 | 0.796 ± 0.139 |
5.482 ± 1.179 | 3.095 ± 0.761 |
6.72 ± 0.694 | 4.951 ± 0.23 |
3.36 ± 0.114 | 6.012 ± 0.746 |
3.36 ± 1.335 | 8.93 ± 0.192 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.653 ± 0.326 | 4.775 ± 1.139 |
6.985 ± 0.445 | 3.89 ± 0.416 |
5.217 ± 0.475 | 7.162 ± 0.326 |
7.25 ± 0.53 | 4.598 ± 0.468 |
2.122 ± 0.503 | 4.332 ± 0.934 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
