
Arenimonas oryziterrae DSM 21050 = YC6267
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; Arenimonas; Arenimonas oryziterrae
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
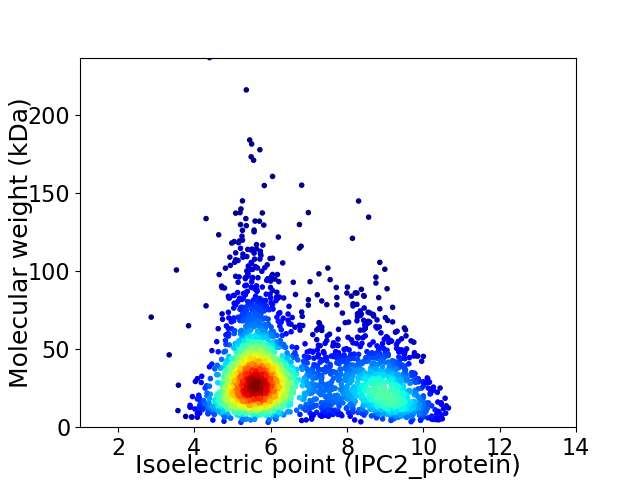
Virtual 2D-PAGE plot for 2897 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
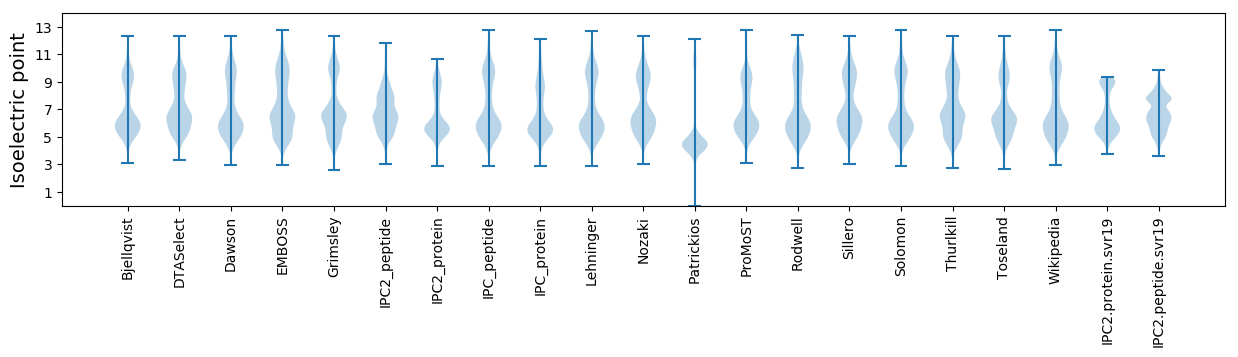
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A091B9K3|A0A091B9K3_9GAMM Signal peptidase I OS=Arenimonas oryziterrae DSM 21050 = YC6267 OX=1121015 GN=N789_04415 PE=3 SV=1
MM1 pKa = 7.27SADD4 pKa = 4.11PRR6 pKa = 11.84PLPPPPPDD14 pKa = 4.03PADD17 pKa = 3.8CCGSGCVRR25 pKa = 11.84CIFDD29 pKa = 5.19LYY31 pKa = 11.35DD32 pKa = 4.25DD33 pKa = 4.17ALARR37 pKa = 11.84YY38 pKa = 7.74DD39 pKa = 3.53AQLAQWLTRR48 pKa = 11.84HH49 pKa = 6.56PDD51 pKa = 2.99AAADD55 pKa = 3.96ADD57 pKa = 4.21SMPP60 pKa = 4.17
MM1 pKa = 7.27SADD4 pKa = 4.11PRR6 pKa = 11.84PLPPPPPDD14 pKa = 4.03PADD17 pKa = 3.8CCGSGCVRR25 pKa = 11.84CIFDD29 pKa = 5.19LYY31 pKa = 11.35DD32 pKa = 4.25DD33 pKa = 4.17ALARR37 pKa = 11.84YY38 pKa = 7.74DD39 pKa = 3.53AQLAQWLTRR48 pKa = 11.84HH49 pKa = 6.56PDD51 pKa = 2.99AAADD55 pKa = 3.96ADD57 pKa = 4.21SMPP60 pKa = 4.17
Molecular weight: 6.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A091AUP7|A0A091AUP7_9GAMM 3-ketoacyl-ACP reductase OS=Arenimonas oryziterrae DSM 21050 = YC6267 OX=1121015 GN=fabG PE=4 SV=1
MM1 pKa = 7.6RR2 pKa = 11.84PVDD5 pKa = 3.69AGAGNGMRR13 pKa = 11.84EE14 pKa = 4.0RR15 pKa = 11.84LNYY18 pKa = 8.72AWRR21 pKa = 11.84VIATGLSFLSFGIGGVILWLLVFPLLSLLVRR52 pKa = 11.84DD53 pKa = 4.67LEE55 pKa = 4.4VRR57 pKa = 11.84SQKK60 pKa = 10.4ARR62 pKa = 11.84AIIHH66 pKa = 6.47RR67 pKa = 11.84SFALFIGFMHH77 pKa = 7.0RR78 pKa = 11.84MGVLTYY84 pKa = 8.79EE85 pKa = 3.62IRR87 pKa = 11.84GAEE90 pKa = 3.8RR91 pKa = 11.84LEE93 pKa = 3.88RR94 pKa = 11.84RR95 pKa = 11.84GLLVLANHH103 pKa = 6.75PTLIDD108 pKa = 3.52VVFLVSLIPNADD120 pKa = 3.56CVVKK124 pKa = 10.75SRR126 pKa = 11.84LATNPFTRR134 pKa = 11.84GPVRR138 pKa = 11.84ATRR141 pKa = 11.84YY142 pKa = 8.86ICNDD146 pKa = 2.82NGAGLIEE153 pKa = 4.63DD154 pKa = 5.51CIASLRR160 pKa = 11.84SGKK163 pKa = 9.65NLIIFPEE170 pKa = 4.75GTRR173 pKa = 11.84TPRR176 pKa = 11.84TGPATLQRR184 pKa = 11.84GAANIAVRR192 pKa = 11.84GLRR195 pKa = 11.84DD196 pKa = 3.38ITPVIIRR203 pKa = 11.84CSPPTLSKK211 pKa = 10.64GEE213 pKa = 3.52KK214 pKa = 8.27WYY216 pKa = 10.4RR217 pKa = 11.84VPSRR221 pKa = 11.84RR222 pKa = 11.84FHH224 pKa = 7.04VSIEE228 pKa = 3.96VRR230 pKa = 11.84EE231 pKa = 4.22DD232 pKa = 3.18MPVAPFLAGTSEE244 pKa = 3.89AQAARR249 pKa = 11.84KK250 pKa = 8.06LTEE253 pKa = 3.92HH254 pKa = 5.86LTGYY258 pKa = 10.61FDD260 pKa = 4.58LEE262 pKa = 4.12RR263 pKa = 11.84PIASAA268 pKa = 3.9
MM1 pKa = 7.6RR2 pKa = 11.84PVDD5 pKa = 3.69AGAGNGMRR13 pKa = 11.84EE14 pKa = 4.0RR15 pKa = 11.84LNYY18 pKa = 8.72AWRR21 pKa = 11.84VIATGLSFLSFGIGGVILWLLVFPLLSLLVRR52 pKa = 11.84DD53 pKa = 4.67LEE55 pKa = 4.4VRR57 pKa = 11.84SQKK60 pKa = 10.4ARR62 pKa = 11.84AIIHH66 pKa = 6.47RR67 pKa = 11.84SFALFIGFMHH77 pKa = 7.0RR78 pKa = 11.84MGVLTYY84 pKa = 8.79EE85 pKa = 3.62IRR87 pKa = 11.84GAEE90 pKa = 3.8RR91 pKa = 11.84LEE93 pKa = 3.88RR94 pKa = 11.84RR95 pKa = 11.84GLLVLANHH103 pKa = 6.75PTLIDD108 pKa = 3.52VVFLVSLIPNADD120 pKa = 3.56CVVKK124 pKa = 10.75SRR126 pKa = 11.84LATNPFTRR134 pKa = 11.84GPVRR138 pKa = 11.84ATRR141 pKa = 11.84YY142 pKa = 8.86ICNDD146 pKa = 2.82NGAGLIEE153 pKa = 4.63DD154 pKa = 5.51CIASLRR160 pKa = 11.84SGKK163 pKa = 9.65NLIIFPEE170 pKa = 4.75GTRR173 pKa = 11.84TPRR176 pKa = 11.84TGPATLQRR184 pKa = 11.84GAANIAVRR192 pKa = 11.84GLRR195 pKa = 11.84DD196 pKa = 3.38ITPVIIRR203 pKa = 11.84CSPPTLSKK211 pKa = 10.64GEE213 pKa = 3.52KK214 pKa = 8.27WYY216 pKa = 10.4RR217 pKa = 11.84VPSRR221 pKa = 11.84RR222 pKa = 11.84FHH224 pKa = 7.04VSIEE228 pKa = 3.96VRR230 pKa = 11.84EE231 pKa = 4.22DD232 pKa = 3.18MPVAPFLAGTSEE244 pKa = 3.89AQAARR249 pKa = 11.84KK250 pKa = 8.06LTEE253 pKa = 3.92HH254 pKa = 5.86LTGYY258 pKa = 10.61FDD260 pKa = 4.58LEE262 pKa = 4.12RR263 pKa = 11.84PIASAA268 pKa = 3.9
Molecular weight: 29.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
942598 |
29 |
2206 |
325.4 |
35.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.663 ± 0.061 | 0.863 ± 0.015 |
5.75 ± 0.035 | 5.421 ± 0.038 |
3.614 ± 0.032 | 8.289 ± 0.044 |
2.17 ± 0.023 | 4.459 ± 0.027 |
3.079 ± 0.037 | 11.033 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.198 ± 0.02 | 2.592 ± 0.032 |
5.287 ± 0.036 | 3.583 ± 0.025 |
7.39 ± 0.046 | 5.274 ± 0.03 |
5.127 ± 0.043 | 7.351 ± 0.036 |
1.491 ± 0.019 | 2.367 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
