
Cyberlindnera jadinii (strain ATCC 18201 / CBS 1600 / BCRC 20928 / JCM 3617 / NBRC 0987 / NRRL Y-1542) (Torula yeast) (Candida utilis)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Phaffomycetaceae; Cyberlindnera; Cyberlindnera jadinii
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
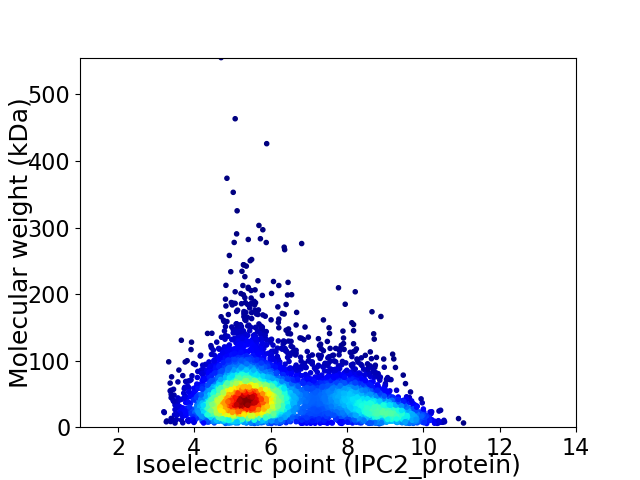
Virtual 2D-PAGE plot for 5994 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E4S1F9|A0A1E4S1F9_CYBJN Asparagine synthase (glutamine-hydrolyzing) OS=Cyberlindnera jadinii (strain ATCC 18201 / CBS 1600 / BCRC 20928 / JCM 3617 / NBRC 0987 / NRRL Y-1542) OX=983966 GN=CYBJADRAFT_173338 PE=4 SV=1
MM1 pKa = 7.84KK2 pKa = 10.5LLQLLASTLCATSLAIADD20 pKa = 3.83IAPMKK25 pKa = 10.45FDD27 pKa = 3.66ILRR30 pKa = 11.84GTDD33 pKa = 3.04MSSAKK38 pKa = 10.04KK39 pKa = 10.17KK40 pKa = 10.0PRR42 pKa = 11.84LLKK45 pKa = 10.14RR46 pKa = 11.84DD47 pKa = 3.22AGDD50 pKa = 3.19IVFSDD55 pKa = 3.21VLYY58 pKa = 10.42QEE60 pKa = 4.43KK61 pKa = 10.53SFYY64 pKa = 10.14SANITIAGTEE74 pKa = 4.14FQVQLDD80 pKa = 4.08TGSSDD85 pKa = 2.84LWVYY89 pKa = 10.63GADD92 pKa = 4.04NPFCASNGGVVVNHH106 pKa = 7.12IDD108 pKa = 3.33QSSTVVTPAEE118 pKa = 3.91ATLDD122 pKa = 3.74CEE124 pKa = 4.55AYY126 pKa = 10.54GIFDD130 pKa = 3.83IDD132 pKa = 3.75NEE134 pKa = 4.33DD135 pKa = 4.72FVTEE139 pKa = 4.14DD140 pKa = 3.08QLLFADD146 pKa = 4.24YY147 pKa = 11.02GDD149 pKa = 3.82GSFAVGVYY157 pKa = 10.13GRR159 pKa = 11.84STVSFGNATLEE170 pKa = 3.94GFMFGVANLSNSSVPVCGLSFEE192 pKa = 4.37VSEE195 pKa = 4.8FSLDD199 pKa = 3.57PEE201 pKa = 4.5MSTDD205 pKa = 3.45SPGGSFMYY213 pKa = 10.56QNLPSAMKK221 pKa = 8.69QTGLIKK227 pKa = 9.91RR228 pKa = 11.84TAFSLWIDD236 pKa = 3.88GEE238 pKa = 4.27SSEE241 pKa = 5.33GYY243 pKa = 10.1ILFGAIDD250 pKa = 3.46HH251 pKa = 6.46SKK253 pKa = 10.97YY254 pKa = 10.57SGEE257 pKa = 4.32LVTLPLLITDD267 pKa = 4.53NSSQPNAYY275 pKa = 9.1QVSLNTVILSDD286 pKa = 3.86GDD288 pKa = 4.08SEE290 pKa = 4.65VDD292 pKa = 3.43VMVNEE297 pKa = 4.29TSVLLDD303 pKa = 3.39SGTSEE308 pKa = 5.17AYY310 pKa = 10.47LPNDD314 pKa = 3.16AVEE317 pKa = 4.79NMAEE321 pKa = 4.03ALSAKK326 pKa = 10.41YY327 pKa = 10.5SDD329 pKa = 4.17DD330 pKa = 3.13QGTYY334 pKa = 10.9VLDD337 pKa = 4.1CPKK340 pKa = 10.42EE341 pKa = 4.18STNITITFTFDD352 pKa = 3.98GIDD355 pKa = 3.1IQIPLEE361 pKa = 4.33SFLSATDD368 pKa = 3.63SSGEE372 pKa = 3.85KK373 pKa = 9.19CALMIYY379 pKa = 10.12PGGGNNILGDD389 pKa = 3.64VFLQNVYY396 pKa = 9.97VVFDD400 pKa = 3.55QDD402 pKa = 3.25GLQARR407 pKa = 11.84IAPVDD412 pKa = 3.87NNNDD416 pKa = 3.52SADD419 pKa = 3.56IEE421 pKa = 4.29ILDD424 pKa = 4.06SVKK427 pKa = 10.19TPSGGYY433 pKa = 9.92NNEE436 pKa = 4.06TLTATASWDD445 pKa = 3.32GTLATTEE452 pKa = 4.15DD453 pKa = 3.46QAITKK458 pKa = 9.96SLSSYY463 pKa = 10.45LSSDD467 pKa = 3.51YY468 pKa = 11.32GLSTSEE474 pKa = 4.45SATASS479 pKa = 3.37
MM1 pKa = 7.84KK2 pKa = 10.5LLQLLASTLCATSLAIADD20 pKa = 3.83IAPMKK25 pKa = 10.45FDD27 pKa = 3.66ILRR30 pKa = 11.84GTDD33 pKa = 3.04MSSAKK38 pKa = 10.04KK39 pKa = 10.17KK40 pKa = 10.0PRR42 pKa = 11.84LLKK45 pKa = 10.14RR46 pKa = 11.84DD47 pKa = 3.22AGDD50 pKa = 3.19IVFSDD55 pKa = 3.21VLYY58 pKa = 10.42QEE60 pKa = 4.43KK61 pKa = 10.53SFYY64 pKa = 10.14SANITIAGTEE74 pKa = 4.14FQVQLDD80 pKa = 4.08TGSSDD85 pKa = 2.84LWVYY89 pKa = 10.63GADD92 pKa = 4.04NPFCASNGGVVVNHH106 pKa = 7.12IDD108 pKa = 3.33QSSTVVTPAEE118 pKa = 3.91ATLDD122 pKa = 3.74CEE124 pKa = 4.55AYY126 pKa = 10.54GIFDD130 pKa = 3.83IDD132 pKa = 3.75NEE134 pKa = 4.33DD135 pKa = 4.72FVTEE139 pKa = 4.14DD140 pKa = 3.08QLLFADD146 pKa = 4.24YY147 pKa = 11.02GDD149 pKa = 3.82GSFAVGVYY157 pKa = 10.13GRR159 pKa = 11.84STVSFGNATLEE170 pKa = 3.94GFMFGVANLSNSSVPVCGLSFEE192 pKa = 4.37VSEE195 pKa = 4.8FSLDD199 pKa = 3.57PEE201 pKa = 4.5MSTDD205 pKa = 3.45SPGGSFMYY213 pKa = 10.56QNLPSAMKK221 pKa = 8.69QTGLIKK227 pKa = 9.91RR228 pKa = 11.84TAFSLWIDD236 pKa = 3.88GEE238 pKa = 4.27SSEE241 pKa = 5.33GYY243 pKa = 10.1ILFGAIDD250 pKa = 3.46HH251 pKa = 6.46SKK253 pKa = 10.97YY254 pKa = 10.57SGEE257 pKa = 4.32LVTLPLLITDD267 pKa = 4.53NSSQPNAYY275 pKa = 9.1QVSLNTVILSDD286 pKa = 3.86GDD288 pKa = 4.08SEE290 pKa = 4.65VDD292 pKa = 3.43VMVNEE297 pKa = 4.29TSVLLDD303 pKa = 3.39SGTSEE308 pKa = 5.17AYY310 pKa = 10.47LPNDD314 pKa = 3.16AVEE317 pKa = 4.79NMAEE321 pKa = 4.03ALSAKK326 pKa = 10.41YY327 pKa = 10.5SDD329 pKa = 4.17DD330 pKa = 3.13QGTYY334 pKa = 10.9VLDD337 pKa = 4.1CPKK340 pKa = 10.42EE341 pKa = 4.18STNITITFTFDD352 pKa = 3.98GIDD355 pKa = 3.1IQIPLEE361 pKa = 4.33SFLSATDD368 pKa = 3.63SSGEE372 pKa = 3.85KK373 pKa = 9.19CALMIYY379 pKa = 10.12PGGGNNILGDD389 pKa = 3.64VFLQNVYY396 pKa = 9.97VVFDD400 pKa = 3.55QDD402 pKa = 3.25GLQARR407 pKa = 11.84IAPVDD412 pKa = 3.87NNNDD416 pKa = 3.52SADD419 pKa = 3.56IEE421 pKa = 4.29ILDD424 pKa = 4.06SVKK427 pKa = 10.19TPSGGYY433 pKa = 9.92NNEE436 pKa = 4.06TLTATASWDD445 pKa = 3.32GTLATTEE452 pKa = 4.15DD453 pKa = 3.46QAITKK458 pKa = 9.96SLSSYY463 pKa = 10.45LSSDD467 pKa = 3.51YY468 pKa = 11.32GLSTSEE474 pKa = 4.45SATASS479 pKa = 3.37
Molecular weight: 51.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E4RWW1|A0A1E4RWW1_CYBJN Zn(2)-C6 fungal-type domain-containing protein OS=Cyberlindnera jadinii (strain ATCC 18201 / CBS 1600 / BCRC 20928 / JCM 3617 / NBRC 0987 / NRRL Y-1542) OX=983966 GN=CYBJADRAFT_164065 PE=4 SV=1
MM1 pKa = 7.94PSQKK5 pKa = 10.34SFKK8 pKa = 9.84TKK10 pKa = 10.18QKK12 pKa = 9.8LAKK15 pKa = 9.58AQRR18 pKa = 11.84TNRR21 pKa = 11.84PLPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TNNTVRR36 pKa = 11.84YY37 pKa = 7.2NAKK40 pKa = 8.87RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.11WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 11.12LNII51 pKa = 3.83
MM1 pKa = 7.94PSQKK5 pKa = 10.34SFKK8 pKa = 9.84TKK10 pKa = 10.18QKK12 pKa = 9.8LAKK15 pKa = 9.58AQRR18 pKa = 11.84TNRR21 pKa = 11.84PLPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TNNTVRR36 pKa = 11.84YY37 pKa = 7.2NAKK40 pKa = 8.87RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.11WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 11.12LNII51 pKa = 3.83
Molecular weight: 6.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2597173 |
50 |
4911 |
433.3 |
48.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.18 ± 0.031 | 1.205 ± 0.011 |
6.115 ± 0.022 | 6.736 ± 0.026 |
4.369 ± 0.022 | 5.588 ± 0.03 |
2.209 ± 0.014 | 5.961 ± 0.022 |
6.577 ± 0.032 | 9.741 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.015 ± 0.013 | 4.66 ± 0.018 |
4.382 ± 0.027 | 4.167 ± 0.03 |
4.628 ± 0.022 | 8.473 ± 0.036 |
5.928 ± 0.03 | 6.614 ± 0.021 |
1.132 ± 0.011 | 3.319 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
