
Leptonychotes weddellii papillomavirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae; non-primate mammal papillomaviruses
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
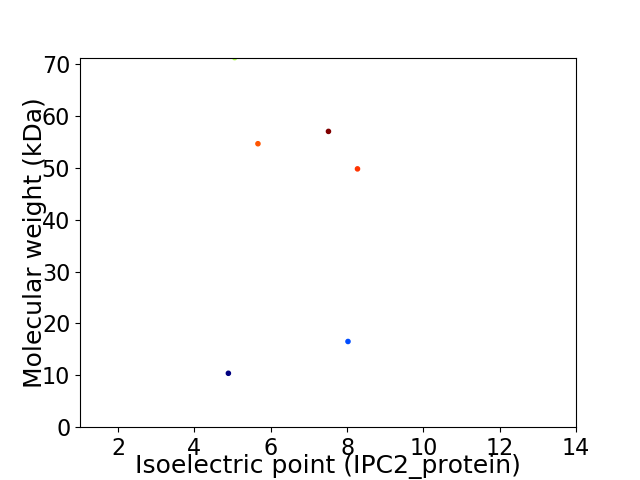
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2I8B2P6|A0A2I8B2P6_9PAPI Replication protein E1 OS=Leptonychotes weddellii papillomavirus 3 OX=2077304 GN=E1 PE=3 SV=1
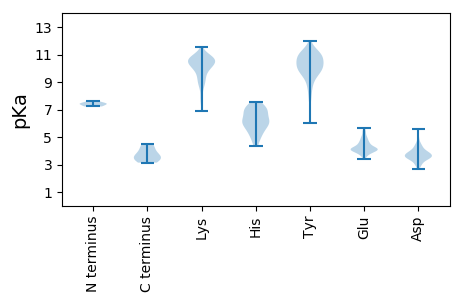
MM1 pKa = 7.61IGTEE5 pKa = 4.13PTLKK9 pKa = 10.68DD10 pKa = 3.16IVLGEE15 pKa = 4.18QPVPADD21 pKa = 4.33DD22 pKa = 3.97LWCYY26 pKa = 10.94EE27 pKa = 4.08EE28 pKa = 5.38LPPEE32 pKa = 3.94EE33 pKa = 5.1AEE35 pKa = 4.89RR36 pKa = 11.84SDD38 pKa = 3.35QAPYY42 pKa = 9.93RR43 pKa = 11.84VHH45 pKa = 7.17ASCGFCGRR53 pKa = 11.84GIEE56 pKa = 4.26LVVLSSRR63 pKa = 11.84VAIRR67 pKa = 11.84TLQHH71 pKa = 6.84LLLEE75 pKa = 4.71NLDD78 pKa = 4.11IVCPDD83 pKa = 3.84CARR86 pKa = 11.84TRR88 pKa = 11.84ALHH91 pKa = 6.35HH92 pKa = 6.63GGG94 pKa = 3.46
MM1 pKa = 7.61IGTEE5 pKa = 4.13PTLKK9 pKa = 10.68DD10 pKa = 3.16IVLGEE15 pKa = 4.18QPVPADD21 pKa = 4.33DD22 pKa = 3.97LWCYY26 pKa = 10.94EE27 pKa = 4.08EE28 pKa = 5.38LPPEE32 pKa = 3.94EE33 pKa = 5.1AEE35 pKa = 4.89RR36 pKa = 11.84SDD38 pKa = 3.35QAPYY42 pKa = 9.93RR43 pKa = 11.84VHH45 pKa = 7.17ASCGFCGRR53 pKa = 11.84GIEE56 pKa = 4.26LVVLSSRR63 pKa = 11.84VAIRR67 pKa = 11.84TLQHH71 pKa = 6.84LLLEE75 pKa = 4.71NLDD78 pKa = 4.11IVCPDD83 pKa = 3.84CARR86 pKa = 11.84TRR88 pKa = 11.84ALHH91 pKa = 6.35HH92 pKa = 6.63GGG94 pKa = 3.46
Molecular weight: 10.39 kDa
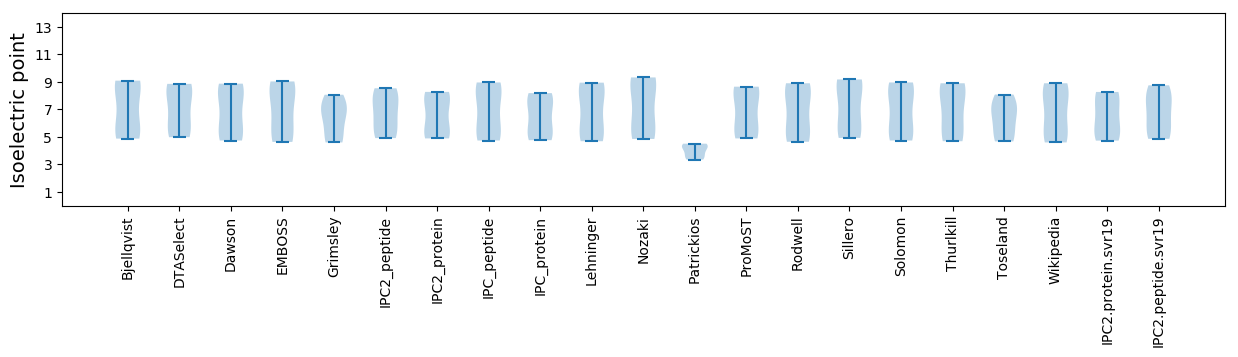
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2I8B2Q2|A0A2I8B2Q2_9PAPI Minor capsid protein L2 OS=Leptonychotes weddellii papillomavirus 3 OX=2077304 GN=L2 PE=3 SV=1
MM1 pKa = 7.41VEE3 pKa = 3.97NLAKK7 pKa = 10.65RR8 pKa = 11.84LDD10 pKa = 3.75LVRR13 pKa = 11.84DD14 pKa = 3.82GLLDD18 pKa = 4.06LYY20 pKa = 10.2EE21 pKa = 4.86RR22 pKa = 11.84CDD24 pKa = 4.53DD25 pKa = 4.02SLEE28 pKa = 4.23SQLKK32 pKa = 8.81HH33 pKa = 4.91WQLLRR38 pKa = 11.84QEE40 pKa = 4.05YY41 pKa = 10.43AILYY45 pKa = 6.48FARR48 pKa = 11.84RR49 pKa = 11.84SGIIRR54 pKa = 11.84VGCQPVPPAQVSRR67 pKa = 11.84EE68 pKa = 3.86RR69 pKa = 11.84ARR71 pKa = 11.84DD72 pKa = 3.72AIEE75 pKa = 3.85VHH77 pKa = 6.95LMLQSLQQSNFCNEE91 pKa = 3.7PWTLADD97 pKa = 3.75TSLEE101 pKa = 4.13RR102 pKa = 11.84YY103 pKa = 7.53KK104 pKa = 10.53TEE106 pKa = 4.97PQGCWKK112 pKa = 10.22KK113 pKa = 9.83NPEE116 pKa = 4.2MVEE119 pKa = 3.58VRR121 pKa = 11.84YY122 pKa = 10.89DD123 pKa = 3.57GDD125 pKa = 3.65RR126 pKa = 11.84EE127 pKa = 4.0NATLHH132 pKa = 5.91TLWQDD137 pKa = 2.71IYY139 pKa = 11.04YY140 pKa = 10.28RR141 pKa = 11.84NSDD144 pKa = 3.81GVWCKK149 pKa = 7.72THH151 pKa = 5.99GQVDD155 pKa = 3.76GHH157 pKa = 6.17GLFYY161 pKa = 10.91LDD163 pKa = 4.39NGCKK167 pKa = 9.56RR168 pKa = 11.84YY169 pKa = 10.03YY170 pKa = 11.01VYY172 pKa = 10.53FADD175 pKa = 4.62DD176 pKa = 3.3AKK178 pKa = 10.7KK179 pKa = 10.51YY180 pKa = 9.54SRR182 pKa = 11.84SGIWEE187 pKa = 3.82VEE189 pKa = 4.17YY190 pKa = 11.01KK191 pKa = 10.91NEE193 pKa = 3.96ILSPLDD199 pKa = 3.47SVTSTTAPAHH209 pKa = 5.22VCRR212 pKa = 11.84PPEE215 pKa = 4.52GPARR219 pKa = 11.84TPARR223 pKa = 11.84GSAGRR228 pKa = 11.84LSTPPEE234 pKa = 4.05PRR236 pKa = 11.84HH237 pKa = 5.71TPWSPQVSTSRR248 pKa = 11.84ADD250 pKa = 3.65VPDD253 pKa = 3.59TTSPLGGPVPALAAQPTLPTAVSRR277 pKa = 11.84SWKK280 pKa = 9.39SATRR284 pKa = 11.84RR285 pKa = 11.84HH286 pKa = 5.74RR287 pKa = 11.84RR288 pKa = 11.84CLGPVTAATPGATDD302 pKa = 3.36TRR304 pKa = 11.84GGAAAAPYY312 pKa = 9.87SAEE315 pKa = 4.56APPSSPTPAEE325 pKa = 4.16GPEE328 pKa = 4.08RR329 pKa = 11.84EE330 pKa = 4.76PEE332 pKa = 4.05RR333 pKa = 11.84QPADD337 pKa = 3.52PGRR340 pKa = 11.84RR341 pKa = 11.84LPPAARR347 pKa = 11.84PNRR350 pKa = 11.84GRR352 pKa = 11.84PGRR355 pKa = 11.84LLRR358 pKa = 11.84GTGDD362 pKa = 3.71CSPALAVLRR371 pKa = 11.84GSGNVLKK378 pKa = 10.53CYY380 pKa = 10.26RR381 pKa = 11.84YY382 pKa = 9.86RR383 pKa = 11.84IKK385 pKa = 10.59KK386 pKa = 7.63KK387 pKa = 9.47HH388 pKa = 5.6RR389 pKa = 11.84RR390 pKa = 11.84LFRR393 pKa = 11.84AVSTTWHH400 pKa = 5.2WTGEE404 pKa = 4.21DD405 pKa = 3.67GADD408 pKa = 3.37RR409 pKa = 11.84AGEE412 pKa = 3.83ARR414 pKa = 11.84LLVWFEE420 pKa = 4.19NPAQRR425 pKa = 11.84SEE427 pKa = 4.23FFEE430 pKa = 4.49SVPMPPSVSAYY441 pKa = 9.82EE442 pKa = 4.15GLGGLL447 pKa = 4.48
MM1 pKa = 7.41VEE3 pKa = 3.97NLAKK7 pKa = 10.65RR8 pKa = 11.84LDD10 pKa = 3.75LVRR13 pKa = 11.84DD14 pKa = 3.82GLLDD18 pKa = 4.06LYY20 pKa = 10.2EE21 pKa = 4.86RR22 pKa = 11.84CDD24 pKa = 4.53DD25 pKa = 4.02SLEE28 pKa = 4.23SQLKK32 pKa = 8.81HH33 pKa = 4.91WQLLRR38 pKa = 11.84QEE40 pKa = 4.05YY41 pKa = 10.43AILYY45 pKa = 6.48FARR48 pKa = 11.84RR49 pKa = 11.84SGIIRR54 pKa = 11.84VGCQPVPPAQVSRR67 pKa = 11.84EE68 pKa = 3.86RR69 pKa = 11.84ARR71 pKa = 11.84DD72 pKa = 3.72AIEE75 pKa = 3.85VHH77 pKa = 6.95LMLQSLQQSNFCNEE91 pKa = 3.7PWTLADD97 pKa = 3.75TSLEE101 pKa = 4.13RR102 pKa = 11.84YY103 pKa = 7.53KK104 pKa = 10.53TEE106 pKa = 4.97PQGCWKK112 pKa = 10.22KK113 pKa = 9.83NPEE116 pKa = 4.2MVEE119 pKa = 3.58VRR121 pKa = 11.84YY122 pKa = 10.89DD123 pKa = 3.57GDD125 pKa = 3.65RR126 pKa = 11.84EE127 pKa = 4.0NATLHH132 pKa = 5.91TLWQDD137 pKa = 2.71IYY139 pKa = 11.04YY140 pKa = 10.28RR141 pKa = 11.84NSDD144 pKa = 3.81GVWCKK149 pKa = 7.72THH151 pKa = 5.99GQVDD155 pKa = 3.76GHH157 pKa = 6.17GLFYY161 pKa = 10.91LDD163 pKa = 4.39NGCKK167 pKa = 9.56RR168 pKa = 11.84YY169 pKa = 10.03YY170 pKa = 11.01VYY172 pKa = 10.53FADD175 pKa = 4.62DD176 pKa = 3.3AKK178 pKa = 10.7KK179 pKa = 10.51YY180 pKa = 9.54SRR182 pKa = 11.84SGIWEE187 pKa = 3.82VEE189 pKa = 4.17YY190 pKa = 11.01KK191 pKa = 10.91NEE193 pKa = 3.96ILSPLDD199 pKa = 3.47SVTSTTAPAHH209 pKa = 5.22VCRR212 pKa = 11.84PPEE215 pKa = 4.52GPARR219 pKa = 11.84TPARR223 pKa = 11.84GSAGRR228 pKa = 11.84LSTPPEE234 pKa = 4.05PRR236 pKa = 11.84HH237 pKa = 5.71TPWSPQVSTSRR248 pKa = 11.84ADD250 pKa = 3.65VPDD253 pKa = 3.59TTSPLGGPVPALAAQPTLPTAVSRR277 pKa = 11.84SWKK280 pKa = 9.39SATRR284 pKa = 11.84RR285 pKa = 11.84HH286 pKa = 5.74RR287 pKa = 11.84RR288 pKa = 11.84CLGPVTAATPGATDD302 pKa = 3.36TRR304 pKa = 11.84GGAAAAPYY312 pKa = 9.87SAEE315 pKa = 4.56APPSSPTPAEE325 pKa = 4.16GPEE328 pKa = 4.08RR329 pKa = 11.84EE330 pKa = 4.76PEE332 pKa = 4.05RR333 pKa = 11.84QPADD337 pKa = 3.52PGRR340 pKa = 11.84RR341 pKa = 11.84LPPAARR347 pKa = 11.84PNRR350 pKa = 11.84GRR352 pKa = 11.84PGRR355 pKa = 11.84LLRR358 pKa = 11.84GTGDD362 pKa = 3.71CSPALAVLRR371 pKa = 11.84GSGNVLKK378 pKa = 10.53CYY380 pKa = 10.26RR381 pKa = 11.84YY382 pKa = 9.86RR383 pKa = 11.84IKK385 pKa = 10.59KK386 pKa = 7.63KK387 pKa = 9.47HH388 pKa = 5.6RR389 pKa = 11.84RR390 pKa = 11.84LFRR393 pKa = 11.84AVSTTWHH400 pKa = 5.2WTGEE404 pKa = 4.21DD405 pKa = 3.67GADD408 pKa = 3.37RR409 pKa = 11.84AGEE412 pKa = 3.83ARR414 pKa = 11.84LLVWFEE420 pKa = 4.19NPAQRR425 pKa = 11.84SEE427 pKa = 4.23FFEE430 pKa = 4.49SVPMPPSVSAYY441 pKa = 9.82EE442 pKa = 4.15GLGGLL447 pKa = 4.48
Molecular weight: 49.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2336 |
94 |
632 |
389.3 |
43.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.449 ± 0.476 | 2.568 ± 0.724 |
5.437 ± 0.21 | 6.507 ± 0.54 |
3.81 ± 0.525 | 7.705 ± 0.758 |
2.226 ± 0.183 | 3.767 ± 0.509 |
3.938 ± 0.649 | 8.69 ± 0.801 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.584 ± 0.524 | 3.682 ± 0.753 |
6.807 ± 1.007 | 3.981 ± 0.408 |
7.363 ± 0.839 | 7.834 ± 0.651 |
5.95 ± 0.477 | 5.95 ± 0.764 |
1.627 ± 0.42 | 3.082 ± 0.325 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
