
Actinomyces denticolens
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Actinomycetales; Actinomycetaceae; Actinomyces
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
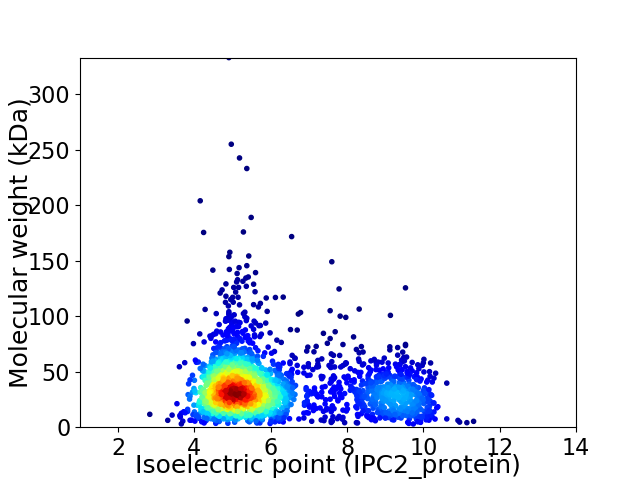
Virtual 2D-PAGE plot for 2279 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M5ZXV7|A0A1M5ZXV7_9ACTO Adenylate cyclase OS=Actinomyces denticolens OX=52767 GN=SAMN05216246_10137 PE=3 SV=1
MM1 pKa = 7.15STGPYY6 pKa = 9.86DD7 pKa = 4.51PNIQYY12 pKa = 10.49DD13 pKa = 3.98AAPASPTGPSTPMGAPAGGPPAYY36 pKa = 10.31GMGGQYY42 pKa = 8.15PAPGQAPPTPPSTKK56 pKa = 9.67RR57 pKa = 11.84KK58 pKa = 10.21GIIALIAGLAVLSLLVTGGILAAYY82 pKa = 8.62KK83 pKa = 10.67DD84 pKa = 4.2SGDD87 pKa = 4.14SSDD90 pKa = 3.7SAAGGSSASTGADD103 pKa = 2.89SSGGKK108 pKa = 10.24LGDD111 pKa = 3.81KK112 pKa = 10.29ASKK115 pKa = 10.35AAVGDD120 pKa = 4.74CIDD123 pKa = 4.22SLTSPSATIVACDD136 pKa = 3.52DD137 pKa = 3.42AGAAFKK143 pKa = 10.84VVAASTTKK151 pKa = 9.77EE152 pKa = 3.85ACATTPGGDD161 pKa = 3.49YY162 pKa = 10.75FDD164 pKa = 6.72DD165 pKa = 3.84STCYY169 pKa = 10.72NDD171 pKa = 4.56LSTGIPVEE179 pKa = 4.08EE180 pKa = 4.48SVNDD184 pKa = 4.01AKK186 pKa = 11.28AGDD189 pKa = 4.47CVAADD194 pKa = 4.66DD195 pKa = 4.7TDD197 pKa = 3.61SSNPSVRR204 pKa = 11.84KK205 pKa = 8.03TDD207 pKa = 3.42CSTTGALRR215 pKa = 11.84VEE217 pKa = 4.55GTLPGVSGVPTIAIPTIDD235 pKa = 3.65VSNPGPAPTLEE246 pKa = 4.24DD247 pKa = 3.29VRR249 pKa = 11.84TQYY252 pKa = 10.96IIDD255 pKa = 3.5QCVAAGSTGTTAVYY269 pKa = 10.06SWNLGGTSLTGFQTDD284 pKa = 3.68TPSSYY289 pKa = 11.33DD290 pKa = 3.26AVLCLSQGAA299 pKa = 3.79
MM1 pKa = 7.15STGPYY6 pKa = 9.86DD7 pKa = 4.51PNIQYY12 pKa = 10.49DD13 pKa = 3.98AAPASPTGPSTPMGAPAGGPPAYY36 pKa = 10.31GMGGQYY42 pKa = 8.15PAPGQAPPTPPSTKK56 pKa = 9.67RR57 pKa = 11.84KK58 pKa = 10.21GIIALIAGLAVLSLLVTGGILAAYY82 pKa = 8.62KK83 pKa = 10.67DD84 pKa = 4.2SGDD87 pKa = 4.14SSDD90 pKa = 3.7SAAGGSSASTGADD103 pKa = 2.89SSGGKK108 pKa = 10.24LGDD111 pKa = 3.81KK112 pKa = 10.29ASKK115 pKa = 10.35AAVGDD120 pKa = 4.74CIDD123 pKa = 4.22SLTSPSATIVACDD136 pKa = 3.52DD137 pKa = 3.42AGAAFKK143 pKa = 10.84VVAASTTKK151 pKa = 9.77EE152 pKa = 3.85ACATTPGGDD161 pKa = 3.49YY162 pKa = 10.75FDD164 pKa = 6.72DD165 pKa = 3.84STCYY169 pKa = 10.72NDD171 pKa = 4.56LSTGIPVEE179 pKa = 4.08EE180 pKa = 4.48SVNDD184 pKa = 4.01AKK186 pKa = 11.28AGDD189 pKa = 4.47CVAADD194 pKa = 4.66DD195 pKa = 4.7TDD197 pKa = 3.61SSNPSVRR204 pKa = 11.84KK205 pKa = 8.03TDD207 pKa = 3.42CSTTGALRR215 pKa = 11.84VEE217 pKa = 4.55GTLPGVSGVPTIAIPTIDD235 pKa = 3.65VSNPGPAPTLEE246 pKa = 4.24DD247 pKa = 3.29VRR249 pKa = 11.84TQYY252 pKa = 10.96IIDD255 pKa = 3.5QCVAAGSTGTTAVYY269 pKa = 10.06SWNLGGTSLTGFQTDD284 pKa = 3.68TPSSYY289 pKa = 11.33DD290 pKa = 3.26AVLCLSQGAA299 pKa = 3.79
Molecular weight: 29.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M6EM70|A0A1M6EM70_9ACTO Cell division protein CrgA OS=Actinomyces denticolens OX=52767 GN=crgA PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.4VHH17 pKa = 5.52GFRR20 pKa = 11.84KK21 pKa = 9.95RR22 pKa = 11.84MSTRR26 pKa = 11.84AGRR29 pKa = 11.84AVLASRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.61GRR40 pKa = 11.84ARR42 pKa = 11.84LAAA45 pKa = 4.44
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 9.4VHH17 pKa = 5.52GFRR20 pKa = 11.84KK21 pKa = 9.95RR22 pKa = 11.84MSTRR26 pKa = 11.84AGRR29 pKa = 11.84AVLASRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.61GRR40 pKa = 11.84ARR42 pKa = 11.84LAAA45 pKa = 4.44
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
815642 |
27 |
3198 |
357.9 |
37.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.736 ± 0.086 | 0.729 ± 0.014 |
5.931 ± 0.047 | 5.616 ± 0.059 |
2.309 ± 0.027 | 9.583 ± 0.049 |
1.954 ± 0.02 | 4.338 ± 0.037 |
1.75 ± 0.038 | 9.985 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.021 ± 0.022 | 1.686 ± 0.029 |
6.074 ± 0.043 | 2.496 ± 0.023 |
7.734 ± 0.054 | 5.992 ± 0.043 |
5.87 ± 0.042 | 7.875 ± 0.046 |
1.471 ± 0.026 | 1.85 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
