
Bowmanella sp. JS7-9
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Bowmanella; unclassified Bowmanella
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
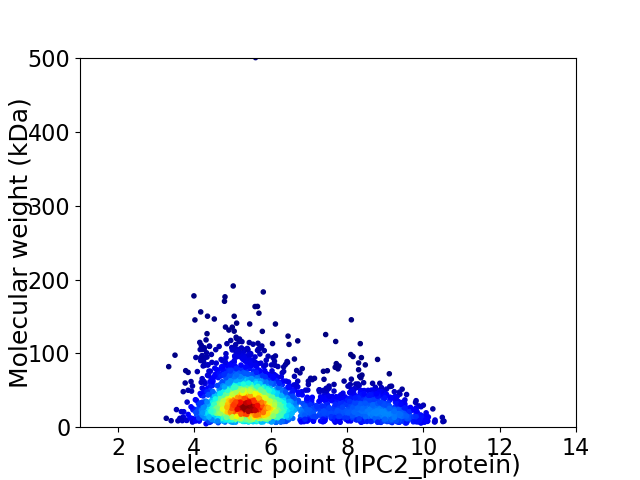
Virtual 2D-PAGE plot for 2974 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q9XH50|A0A4Q9XH50_9ALTE Metal ABC transporter permease OS=Bowmanella sp. JS7-9 OX=1605368 GN=TK45_15905 PE=4 SV=1
MM1 pKa = 6.68NHH3 pKa = 6.79KK4 pKa = 10.46LITVSLIAAGVLAGCGGSNDD24 pKa = 4.51KK25 pKa = 10.58PLEE28 pKa = 4.12QPNPTFALAISDD40 pKa = 4.38APVDD44 pKa = 3.9DD45 pKa = 4.32LSEE48 pKa = 4.17VVVCFNAVEE57 pKa = 4.33LKK59 pKa = 10.67HH60 pKa = 6.91ADD62 pKa = 3.29GSASDD67 pKa = 3.5STFTVGTDD75 pKa = 3.51PQMLTANDD83 pKa = 3.32VCKK86 pKa = 10.79NDD88 pKa = 3.65AGEE91 pKa = 4.39VIPATVGINLLDD103 pKa = 3.75YY104 pKa = 10.44MGMASEE110 pKa = 4.55SLLSEE115 pKa = 4.17VSIAPGTYY123 pKa = 8.49NQLRR127 pKa = 11.84LVMADD132 pKa = 3.25GSYY135 pKa = 9.82GTVKK139 pKa = 9.6EE140 pKa = 4.35TGEE143 pKa = 4.52KK144 pKa = 9.35IDD146 pKa = 3.46VSVPSNEE153 pKa = 4.01LKK155 pKa = 10.88LDD157 pKa = 3.74GFTAALGNTLSFTLEE172 pKa = 3.42FDD174 pKa = 3.67LRR176 pKa = 11.84KK177 pKa = 10.55AMTNPVGQSGYY188 pKa = 8.38FLKK191 pKa = 10.7PRR193 pKa = 11.84GVRR196 pKa = 11.84LVDD199 pKa = 3.39NNQIGHH205 pKa = 4.92VQGTVDD211 pKa = 3.61EE212 pKa = 5.08TILVNNSCTVAPSDD226 pKa = 2.93ITTPVATVYY235 pKa = 10.54LYY237 pKa = 10.67QGVSLDD243 pKa = 4.07SNTLADD249 pKa = 3.5NGGAEE254 pKa = 4.2GVEE257 pKa = 4.4PFASTTVTFDD267 pKa = 2.97GAATYY272 pKa = 9.89SYY274 pKa = 10.72EE275 pKa = 3.88IGYY278 pKa = 9.4VPAGDD283 pKa = 3.77YY284 pKa = 9.26TLSLSCDD291 pKa = 3.4TVDD294 pKa = 5.85DD295 pKa = 4.85PEE297 pKa = 6.71ADD299 pKa = 4.0DD300 pKa = 7.01DD301 pKa = 4.17ITQLQQAEE309 pKa = 4.43VSVSANSTPVAVNFTAGG326 pKa = 3.08
MM1 pKa = 6.68NHH3 pKa = 6.79KK4 pKa = 10.46LITVSLIAAGVLAGCGGSNDD24 pKa = 4.51KK25 pKa = 10.58PLEE28 pKa = 4.12QPNPTFALAISDD40 pKa = 4.38APVDD44 pKa = 3.9DD45 pKa = 4.32LSEE48 pKa = 4.17VVVCFNAVEE57 pKa = 4.33LKK59 pKa = 10.67HH60 pKa = 6.91ADD62 pKa = 3.29GSASDD67 pKa = 3.5STFTVGTDD75 pKa = 3.51PQMLTANDD83 pKa = 3.32VCKK86 pKa = 10.79NDD88 pKa = 3.65AGEE91 pKa = 4.39VIPATVGINLLDD103 pKa = 3.75YY104 pKa = 10.44MGMASEE110 pKa = 4.55SLLSEE115 pKa = 4.17VSIAPGTYY123 pKa = 8.49NQLRR127 pKa = 11.84LVMADD132 pKa = 3.25GSYY135 pKa = 9.82GTVKK139 pKa = 9.6EE140 pKa = 4.35TGEE143 pKa = 4.52KK144 pKa = 9.35IDD146 pKa = 3.46VSVPSNEE153 pKa = 4.01LKK155 pKa = 10.88LDD157 pKa = 3.74GFTAALGNTLSFTLEE172 pKa = 3.42FDD174 pKa = 3.67LRR176 pKa = 11.84KK177 pKa = 10.55AMTNPVGQSGYY188 pKa = 8.38FLKK191 pKa = 10.7PRR193 pKa = 11.84GVRR196 pKa = 11.84LVDD199 pKa = 3.39NNQIGHH205 pKa = 4.92VQGTVDD211 pKa = 3.61EE212 pKa = 5.08TILVNNSCTVAPSDD226 pKa = 2.93ITTPVATVYY235 pKa = 10.54LYY237 pKa = 10.67QGVSLDD243 pKa = 4.07SNTLADD249 pKa = 3.5NGGAEE254 pKa = 4.2GVEE257 pKa = 4.4PFASTTVTFDD267 pKa = 2.97GAATYY272 pKa = 9.89SYY274 pKa = 10.72EE275 pKa = 3.88IGYY278 pKa = 9.4VPAGDD283 pKa = 3.77YY284 pKa = 9.26TLSLSCDD291 pKa = 3.4TVDD294 pKa = 5.85DD295 pKa = 4.85PEE297 pKa = 6.71ADD299 pKa = 4.0DD300 pKa = 7.01DD301 pKa = 4.17ITQLQQAEE309 pKa = 4.43VSVSANSTPVAVNFTAGG326 pKa = 3.08
Molecular weight: 33.99 kDa
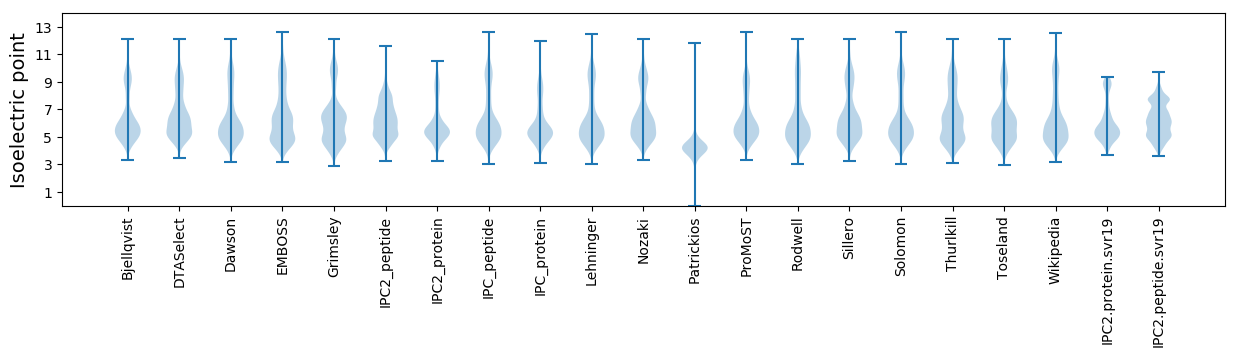
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q9XT28|A0A4Q9XT28_9ALTE Uncharacterized protein OS=Bowmanella sp. JS7-9 OX=1605368 GN=TK45_01415 PE=4 SV=1
MM1 pKa = 7.0MWWLMICIVFAFGVEE16 pKa = 3.75AMTGFGSIVLALSLGALVYY35 pKa = 10.08SIPEE39 pKa = 4.25LVTLLVPLNLVMTLPLCIRR58 pKa = 11.84LRR60 pKa = 11.84QHH62 pKa = 5.25IQWRR66 pKa = 11.84LLLTRR71 pKa = 11.84LLPLMAVGTLLGQALLPWVNPDD93 pKa = 2.71WLKK96 pKa = 11.13LVFAMLITWFALRR109 pKa = 11.84ALLKK113 pKa = 10.36SQALALPTGIANGLITLAGVTHH135 pKa = 6.9GLFASGGPLLVYY147 pKa = 10.73ALARR151 pKa = 11.84SQLPKK156 pKa = 10.24AQFRR160 pKa = 11.84ATLLFVWLCLNSVLSLWFLFSGRR183 pKa = 11.84FEE185 pKa = 4.5GQWPALLQLVPAVLLGMWLGNRR207 pKa = 11.84LHH209 pKa = 6.83HH210 pKa = 7.04KK211 pKa = 10.02IDD213 pKa = 3.35AQRR216 pKa = 11.84FLPSVFAILLLVSVILLVTSLQQVAGG242 pKa = 3.85
MM1 pKa = 7.0MWWLMICIVFAFGVEE16 pKa = 3.75AMTGFGSIVLALSLGALVYY35 pKa = 10.08SIPEE39 pKa = 4.25LVTLLVPLNLVMTLPLCIRR58 pKa = 11.84LRR60 pKa = 11.84QHH62 pKa = 5.25IQWRR66 pKa = 11.84LLLTRR71 pKa = 11.84LLPLMAVGTLLGQALLPWVNPDD93 pKa = 2.71WLKK96 pKa = 11.13LVFAMLITWFALRR109 pKa = 11.84ALLKK113 pKa = 10.36SQALALPTGIANGLITLAGVTHH135 pKa = 6.9GLFASGGPLLVYY147 pKa = 10.73ALARR151 pKa = 11.84SQLPKK156 pKa = 10.24AQFRR160 pKa = 11.84ATLLFVWLCLNSVLSLWFLFSGRR183 pKa = 11.84FEE185 pKa = 4.5GQWPALLQLVPAVLLGMWLGNRR207 pKa = 11.84LHH209 pKa = 6.83HH210 pKa = 7.04KK211 pKa = 10.02IDD213 pKa = 3.35AQRR216 pKa = 11.84FLPSVFAILLLVSVILLVTSLQQVAGG242 pKa = 3.85
Molecular weight: 26.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
973382 |
38 |
4501 |
327.3 |
36.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.718 ± 0.047 | 1.0 ± 0.017 |
5.815 ± 0.037 | 5.652 ± 0.04 |
4.027 ± 0.025 | 7.01 ± 0.041 |
2.346 ± 0.025 | 5.751 ± 0.033 |
4.372 ± 0.042 | 10.711 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.654 ± 0.023 | 3.956 ± 0.03 |
4.097 ± 0.024 | 5.354 ± 0.048 |
5.126 ± 0.035 | 6.098 ± 0.036 |
5.048 ± 0.038 | 6.956 ± 0.04 |
1.325 ± 0.018 | 2.984 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
