
Planktothrix agardhii NIVA-CYA 126/8
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Oscillatoriales; Microcoleaceae; Planktothrix; Planktothrix agardhii
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
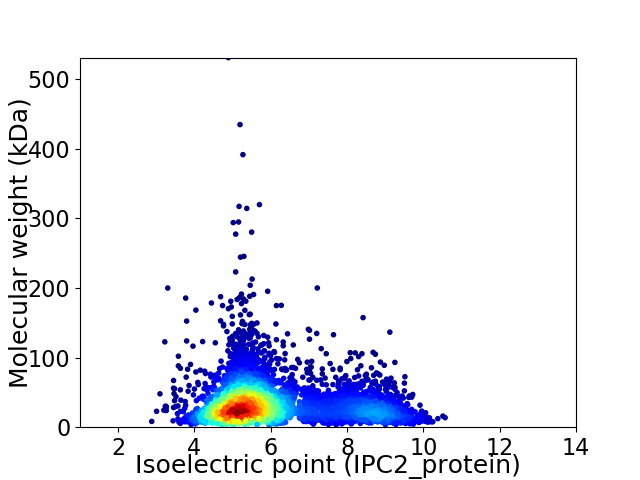
Virtual 2D-PAGE plot for 4186 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A073CS64|A0A073CS64_PLAAG Uma2 domain-containing protein OS=Planktothrix agardhii NIVA-CYA 126/8 OX=388467 GN=A19Y_1855 PE=4 SV=1
MM1 pKa = 7.86IDD3 pKa = 3.05TGVDD7 pKa = 3.48YY8 pKa = 11.13NHH10 pKa = 7.86PDD12 pKa = 3.2LSANIWTNPGEE23 pKa = 4.15IAGDD27 pKa = 4.26GIDD30 pKa = 4.09NDD32 pKa = 4.3SNGYY36 pKa = 9.52IDD38 pKa = 6.35DD39 pKa = 3.64VRR41 pKa = 11.84GWDD44 pKa = 4.12FAYY47 pKa = 11.06NDD49 pKa = 4.04NNPMDD54 pKa = 3.75VHH56 pKa = 6.17GHH58 pKa = 4.65GTHH61 pKa = 5.77VAGTIAGKK69 pKa = 10.46GNNGVGVTGVAWNAKK84 pKa = 8.92IMPLKK89 pKa = 10.31FLNDD93 pKa = 3.77SGWGYY98 pKa = 9.31TSDD101 pKa = 4.48AISAINYY108 pKa = 7.05ATAKK112 pKa = 9.97GVKK115 pKa = 8.44LTNNSWGGDD124 pKa = 3.48GYY126 pKa = 8.95TQALYY131 pKa = 10.77DD132 pKa = 4.8AINTAGQQGALFIAAAGNSSQNTDD156 pKa = 3.12TTPAYY161 pKa = 8.05PASYY165 pKa = 10.52DD166 pKa = 3.45LANIISVASTTRR178 pKa = 11.84TDD180 pKa = 3.34GLSWFSNYY188 pKa = 9.78GATTVDD194 pKa = 3.78LGAPGSDD201 pKa = 4.69IYY203 pKa = 11.2SAWPNSTYY211 pKa = 10.03NTISGTSMASPHH223 pKa = 4.84VTGAAALLWSQNPTWTAQQVKK244 pKa = 9.5NRR246 pKa = 11.84LMSTGDD252 pKa = 3.92SISALNGKK260 pKa = 6.36TVSGKK265 pKa = 10.11RR266 pKa = 11.84LNIYY270 pKa = 9.14NALMSSNLPTVTVNVSPVTVQEE292 pKa = 4.68DD293 pKa = 4.11GSGNLTYY300 pKa = 10.83SFSRR304 pKa = 11.84SGNLTSAMTVNFGVAGTANAAAVGSDD330 pKa = 3.14PADD333 pKa = 3.39YY334 pKa = 10.1TVLTNSAVTFSPSTKK349 pKa = 9.75TGTITFAAGSSTAQLVVDD367 pKa = 4.64PTADD371 pKa = 3.31TLAEE375 pKa = 4.26SQNEE379 pKa = 4.0TVVFNINSGTGYY391 pKa = 10.33IGGTPNTATGTIVSEE406 pKa = 4.03EE407 pKa = 3.96VLPIFTNPNSITIPSSGSASPYY429 pKa = 9.02PSTINVSGVSGNIANIQVSLSGLSHH454 pKa = 6.84TWPDD458 pKa = 3.69DD459 pKa = 3.04VDD461 pKa = 3.54MFLRR465 pKa = 11.84GPGGQKK471 pKa = 10.42VMLMSDD477 pKa = 3.22AGDD480 pKa = 3.49FADD483 pKa = 4.92LNNVNLTFSDD493 pKa = 4.07SASGTLPDD501 pKa = 4.54GSQITSGTYY510 pKa = 9.82RR511 pKa = 11.84PTDD514 pKa = 3.6YY515 pKa = 10.91QVGDD519 pKa = 4.12TFPTPAPAGPYY530 pKa = 8.21GTALSAFNGTNPNGAWQLFVQDD552 pKa = 3.58DD553 pKa = 4.21TGWDD557 pKa = 3.45SGSIAGGWSLTIQRR571 pKa = 11.84TSTINGTAGADD582 pKa = 3.73NLIGTANPDD591 pKa = 3.91IINGLAGNDD600 pKa = 3.72TLNGNTGADD609 pKa = 3.58TLVGGLGNDD618 pKa = 3.01IYY620 pKa = 11.73VVDD623 pKa = 4.03NTGDD627 pKa = 3.29IATEE631 pKa = 4.01LASQGTDD638 pKa = 4.11LIQSSVTYY646 pKa = 8.07TLPANVEE653 pKa = 4.22DD654 pKa = 4.1LTLTGTTAINGTGNALANIITGNTANNILDD684 pKa = 4.01GSSGADD690 pKa = 3.17QLKK693 pKa = 10.74GGTGNDD699 pKa = 3.36TYY701 pKa = 11.75VVDD704 pKa = 3.68NTGDD708 pKa = 3.54VVTEE712 pKa = 4.42LASQGTDD719 pKa = 4.11LIQSSVTYY727 pKa = 8.07TLPANVEE734 pKa = 4.22DD735 pKa = 4.1LTLTGTTAINGTGNTVANIITGNTANNILDD765 pKa = 4.01GSSGADD771 pKa = 3.17QLKK774 pKa = 10.74GGTGNDD780 pKa = 3.36TYY782 pKa = 11.75VVDD785 pKa = 3.68NTGDD789 pKa = 3.54VVTEE793 pKa = 4.42LASQGTDD800 pKa = 4.11LIQSSVTYY808 pKa = 8.07TLPANVEE815 pKa = 4.22DD816 pKa = 4.1LTLTGTTAINGTGNTLANTVTGNTANNILNGGTGNDD852 pKa = 3.47NLIGGSGTDD861 pKa = 3.31QLLGSDD867 pKa = 4.59GNDD870 pKa = 3.42SLSGDD875 pKa = 3.94AGNDD879 pKa = 3.36TLTGGLGADD888 pKa = 3.14KK889 pKa = 10.75FIYY892 pKa = 8.36NTNAAFTTTAVGVDD906 pKa = 4.14TITDD910 pKa = 3.82FNISQTDD917 pKa = 3.49QIVLDD922 pKa = 3.81KK923 pKa = 9.4TTFTSISSAAGTGFSVASEE942 pKa = 4.15FAKK945 pKa = 9.75VTSDD949 pKa = 3.32ALAATSAADD958 pKa = 3.27IVYY961 pKa = 8.13NTATGGLFYY970 pKa = 10.7NQNGTAAGLGTGAQFLTLTNKK991 pKa = 9.62PALTATQFLIQAA1003 pKa = 4.57
MM1 pKa = 7.86IDD3 pKa = 3.05TGVDD7 pKa = 3.48YY8 pKa = 11.13NHH10 pKa = 7.86PDD12 pKa = 3.2LSANIWTNPGEE23 pKa = 4.15IAGDD27 pKa = 4.26GIDD30 pKa = 4.09NDD32 pKa = 4.3SNGYY36 pKa = 9.52IDD38 pKa = 6.35DD39 pKa = 3.64VRR41 pKa = 11.84GWDD44 pKa = 4.12FAYY47 pKa = 11.06NDD49 pKa = 4.04NNPMDD54 pKa = 3.75VHH56 pKa = 6.17GHH58 pKa = 4.65GTHH61 pKa = 5.77VAGTIAGKK69 pKa = 10.46GNNGVGVTGVAWNAKK84 pKa = 8.92IMPLKK89 pKa = 10.31FLNDD93 pKa = 3.77SGWGYY98 pKa = 9.31TSDD101 pKa = 4.48AISAINYY108 pKa = 7.05ATAKK112 pKa = 9.97GVKK115 pKa = 8.44LTNNSWGGDD124 pKa = 3.48GYY126 pKa = 8.95TQALYY131 pKa = 10.77DD132 pKa = 4.8AINTAGQQGALFIAAAGNSSQNTDD156 pKa = 3.12TTPAYY161 pKa = 8.05PASYY165 pKa = 10.52DD166 pKa = 3.45LANIISVASTTRR178 pKa = 11.84TDD180 pKa = 3.34GLSWFSNYY188 pKa = 9.78GATTVDD194 pKa = 3.78LGAPGSDD201 pKa = 4.69IYY203 pKa = 11.2SAWPNSTYY211 pKa = 10.03NTISGTSMASPHH223 pKa = 4.84VTGAAALLWSQNPTWTAQQVKK244 pKa = 9.5NRR246 pKa = 11.84LMSTGDD252 pKa = 3.92SISALNGKK260 pKa = 6.36TVSGKK265 pKa = 10.11RR266 pKa = 11.84LNIYY270 pKa = 9.14NALMSSNLPTVTVNVSPVTVQEE292 pKa = 4.68DD293 pKa = 4.11GSGNLTYY300 pKa = 10.83SFSRR304 pKa = 11.84SGNLTSAMTVNFGVAGTANAAAVGSDD330 pKa = 3.14PADD333 pKa = 3.39YY334 pKa = 10.1TVLTNSAVTFSPSTKK349 pKa = 9.75TGTITFAAGSSTAQLVVDD367 pKa = 4.64PTADD371 pKa = 3.31TLAEE375 pKa = 4.26SQNEE379 pKa = 4.0TVVFNINSGTGYY391 pKa = 10.33IGGTPNTATGTIVSEE406 pKa = 4.03EE407 pKa = 3.96VLPIFTNPNSITIPSSGSASPYY429 pKa = 9.02PSTINVSGVSGNIANIQVSLSGLSHH454 pKa = 6.84TWPDD458 pKa = 3.69DD459 pKa = 3.04VDD461 pKa = 3.54MFLRR465 pKa = 11.84GPGGQKK471 pKa = 10.42VMLMSDD477 pKa = 3.22AGDD480 pKa = 3.49FADD483 pKa = 4.92LNNVNLTFSDD493 pKa = 4.07SASGTLPDD501 pKa = 4.54GSQITSGTYY510 pKa = 9.82RR511 pKa = 11.84PTDD514 pKa = 3.6YY515 pKa = 10.91QVGDD519 pKa = 4.12TFPTPAPAGPYY530 pKa = 8.21GTALSAFNGTNPNGAWQLFVQDD552 pKa = 3.58DD553 pKa = 4.21TGWDD557 pKa = 3.45SGSIAGGWSLTIQRR571 pKa = 11.84TSTINGTAGADD582 pKa = 3.73NLIGTANPDD591 pKa = 3.91IINGLAGNDD600 pKa = 3.72TLNGNTGADD609 pKa = 3.58TLVGGLGNDD618 pKa = 3.01IYY620 pKa = 11.73VVDD623 pKa = 4.03NTGDD627 pKa = 3.29IATEE631 pKa = 4.01LASQGTDD638 pKa = 4.11LIQSSVTYY646 pKa = 8.07TLPANVEE653 pKa = 4.22DD654 pKa = 4.1LTLTGTTAINGTGNALANIITGNTANNILDD684 pKa = 4.01GSSGADD690 pKa = 3.17QLKK693 pKa = 10.74GGTGNDD699 pKa = 3.36TYY701 pKa = 11.75VVDD704 pKa = 3.68NTGDD708 pKa = 3.54VVTEE712 pKa = 4.42LASQGTDD719 pKa = 4.11LIQSSVTYY727 pKa = 8.07TLPANVEE734 pKa = 4.22DD735 pKa = 4.1LTLTGTTAINGTGNTVANIITGNTANNILDD765 pKa = 4.01GSSGADD771 pKa = 3.17QLKK774 pKa = 10.74GGTGNDD780 pKa = 3.36TYY782 pKa = 11.75VVDD785 pKa = 3.68NTGDD789 pKa = 3.54VVTEE793 pKa = 4.42LASQGTDD800 pKa = 4.11LIQSSVTYY808 pKa = 8.07TLPANVEE815 pKa = 4.22DD816 pKa = 4.1LTLTGTTAINGTGNTLANTVTGNTANNILNGGTGNDD852 pKa = 3.47NLIGGSGTDD861 pKa = 3.31QLLGSDD867 pKa = 4.59GNDD870 pKa = 3.42SLSGDD875 pKa = 3.94AGNDD879 pKa = 3.36TLTGGLGADD888 pKa = 3.14KK889 pKa = 10.75FIYY892 pKa = 8.36NTNAAFTTTAVGVDD906 pKa = 4.14TITDD910 pKa = 3.82FNISQTDD917 pKa = 3.49QIVLDD922 pKa = 3.81KK923 pKa = 9.4TTFTSISSAAGTGFSVASEE942 pKa = 4.15FAKK945 pKa = 9.75VTSDD949 pKa = 3.32ALAATSAADD958 pKa = 3.27IVYY961 pKa = 8.13NTATGGLFYY970 pKa = 10.7NQNGTAAGLGTGAQFLTLTNKK991 pKa = 9.62PALTATQFLIQAA1003 pKa = 4.57
Molecular weight: 101.89 kDa
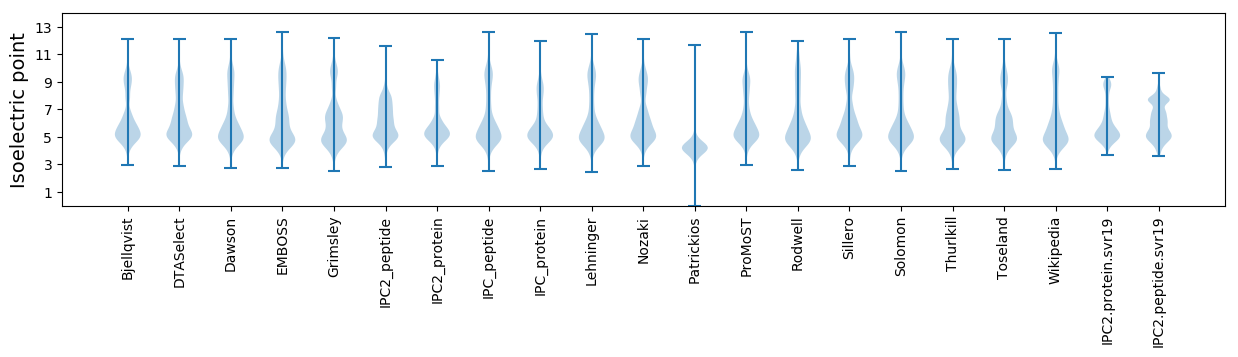
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A073CMB1|A0A073CMB1_PLAAG Isoamyl acetate-hydrolyzing esterase 1 OS=Planktothrix agardhii NIVA-CYA 126/8 OX=388467 GN=A19Y_4130 PE=4 SV=1
MM1 pKa = 7.64EE2 pKa = 5.6KK3 pKa = 10.02SQKK6 pKa = 9.93IKK8 pKa = 11.14LDD10 pKa = 3.24MNRR13 pKa = 11.84NKK15 pKa = 10.44KK16 pKa = 7.37FTKK19 pKa = 10.3GATLFLTGLATVGLVLGEE37 pKa = 4.48AVFSSASAAQQLFCNGRR54 pKa = 11.84MNNGWTYY61 pKa = 11.11SAQFLNGRR69 pKa = 11.84FTEE72 pKa = 4.25IRR74 pKa = 11.84WEE76 pKa = 4.1RR77 pKa = 11.84SGQPPIVSNLSFQSTNNARR96 pKa = 11.84QPVYY100 pKa = 9.67TGSIMAATMVTLVDD114 pKa = 4.15LSRR117 pKa = 11.84GNVRR121 pKa = 11.84PGSQISVTAEE131 pKa = 3.32EE132 pKa = 4.9WGTSTGNCGLSSGNSMGNSSDD153 pKa = 2.68WFTNARR159 pKa = 11.84KK160 pKa = 9.58NLIGSNEE167 pKa = 3.82NQARR171 pKa = 11.84RR172 pKa = 11.84WLTRR176 pKa = 11.84NNFVFNQTMEE186 pKa = 4.02HH187 pKa = 5.15TNRR190 pKa = 11.84RR191 pKa = 11.84IIEE194 pKa = 3.81RR195 pKa = 11.84WSRR198 pKa = 11.84NNPNGVIDD206 pKa = 4.46VIFVNNRR213 pKa = 11.84VSDD216 pKa = 3.9VVMVRR221 pKa = 3.56
MM1 pKa = 7.64EE2 pKa = 5.6KK3 pKa = 10.02SQKK6 pKa = 9.93IKK8 pKa = 11.14LDD10 pKa = 3.24MNRR13 pKa = 11.84NKK15 pKa = 10.44KK16 pKa = 7.37FTKK19 pKa = 10.3GATLFLTGLATVGLVLGEE37 pKa = 4.48AVFSSASAAQQLFCNGRR54 pKa = 11.84MNNGWTYY61 pKa = 11.11SAQFLNGRR69 pKa = 11.84FTEE72 pKa = 4.25IRR74 pKa = 11.84WEE76 pKa = 4.1RR77 pKa = 11.84SGQPPIVSNLSFQSTNNARR96 pKa = 11.84QPVYY100 pKa = 9.67TGSIMAATMVTLVDD114 pKa = 4.15LSRR117 pKa = 11.84GNVRR121 pKa = 11.84PGSQISVTAEE131 pKa = 3.32EE132 pKa = 4.9WGTSTGNCGLSSGNSMGNSSDD153 pKa = 2.68WFTNARR159 pKa = 11.84KK160 pKa = 9.58NLIGSNEE167 pKa = 3.82NQARR171 pKa = 11.84RR172 pKa = 11.84WLTRR176 pKa = 11.84NNFVFNQTMEE186 pKa = 4.02HH187 pKa = 5.15TNRR190 pKa = 11.84RR191 pKa = 11.84IIEE194 pKa = 3.81RR195 pKa = 11.84WSRR198 pKa = 11.84NNPNGVIDD206 pKa = 4.46VIFVNNRR213 pKa = 11.84VSDD216 pKa = 3.9VVMVRR221 pKa = 3.56
Molecular weight: 24.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1394687 |
28 |
4714 |
333.2 |
37.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.87 ± 0.034 | 0.968 ± 0.011 |
4.985 ± 0.028 | 6.498 ± 0.039 |
4.05 ± 0.026 | 6.548 ± 0.04 |
1.751 ± 0.016 | 7.499 ± 0.033 |
5.131 ± 0.032 | 11.151 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.816 ± 0.017 | 4.783 ± 0.035 |
4.921 ± 0.031 | 5.678 ± 0.036 |
4.594 ± 0.028 | 6.408 ± 0.031 |
5.686 ± 0.033 | 6.086 ± 0.032 |
1.407 ± 0.015 | 3.169 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
