
Ornithinimicrobium sp. H23M54
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Ornithinimicrobiaceae; Ornithinimicrobium; unclassified Ornithinimicrobium
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
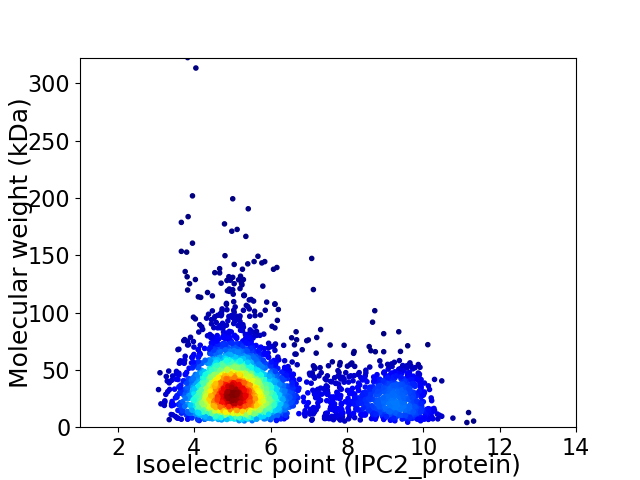
Virtual 2D-PAGE plot for 3622 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
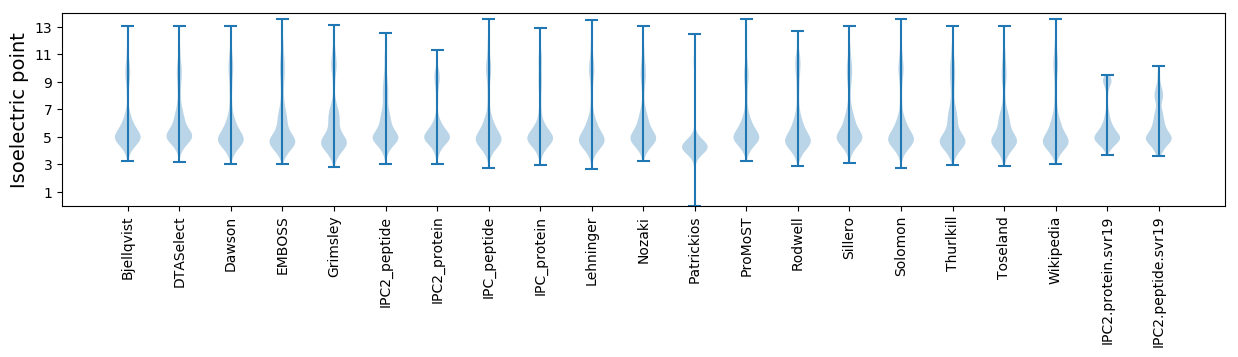
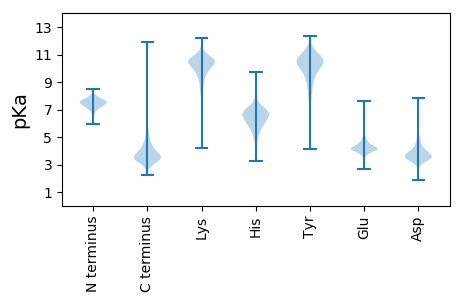
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A516GEQ0|A0A516GEQ0_9MICO Glycosyltransferase family 2 protein OS=Ornithinimicrobium sp. H23M54 OX=2594265 GN=FNH13_18165 PE=4 SV=1
MM1 pKa = 7.5AVHH4 pKa = 6.16GHH6 pKa = 5.92RR7 pKa = 11.84ARR9 pKa = 11.84RR10 pKa = 11.84WLAAVAGASLVASGFGLAGASAQPPQPMAEE40 pKa = 3.86VGATDD45 pKa = 4.83KK46 pKa = 10.92IDD48 pKa = 3.8EE49 pKa = 4.35EE50 pKa = 4.67VSAQIEE56 pKa = 4.4EE57 pKa = 4.23KK58 pKa = 10.64GAADD62 pKa = 3.71FWVRR66 pKa = 11.84FDD68 pKa = 4.24GRR70 pKa = 11.84PDD72 pKa = 3.1MSQFSSISDD81 pKa = 3.05WDD83 pKa = 3.51ARR85 pKa = 11.84GQAVYY90 pKa = 10.66DD91 pKa = 4.13ALTEE95 pKa = 4.16SADD98 pKa = 4.32ASQQDD103 pKa = 3.16IRR105 pKa = 11.84EE106 pKa = 4.04QLDD109 pKa = 3.47ADD111 pKa = 3.95GVDD114 pKa = 4.4YY115 pKa = 11.03QAFWATNAIRR125 pKa = 11.84ITSGDD130 pKa = 3.3QSLVSTLAADD140 pKa = 3.62SGVEE144 pKa = 4.32GIYY147 pKa = 8.5PTTTYY152 pKa = 8.86EE153 pKa = 4.0APEE156 pKa = 3.97LVEE159 pKa = 4.92ADD161 pKa = 4.03RR162 pKa = 11.84EE163 pKa = 4.23MSPSAVEE170 pKa = 3.77WGVADD175 pKa = 4.75INADD179 pKa = 4.1DD180 pKa = 3.43VWSTYY185 pKa = 9.22GVHH188 pKa = 6.84GEE190 pKa = 4.35GIVVGSIDD198 pKa = 4.3TGVQWDD204 pKa = 4.19HH205 pKa = 6.93PALVDD210 pKa = 4.72AYY212 pKa = 10.43RR213 pKa = 11.84GNNGDD218 pKa = 3.42GTFTHH223 pKa = 7.61DD224 pKa = 3.92YY225 pKa = 9.95NWYY228 pKa = 9.31DD229 pKa = 3.43AAGTSPEE236 pKa = 4.22VPADD240 pKa = 3.81GNGHH244 pKa = 4.99GTHH247 pKa = 5.45VTGTMVGDD255 pKa = 4.21DD256 pKa = 3.53GGEE259 pKa = 4.11NQVGVAPGATWISANGCCPSDD280 pKa = 3.48QALIDD285 pKa = 3.6SGQWMLAPTKK295 pKa = 10.68LDD297 pKa = 3.24GTSPDD302 pKa = 3.36TTKK305 pKa = 10.66RR306 pKa = 11.84PHH308 pKa = 7.13IINNSWGTTLPSNDD322 pKa = 3.57PFMEE326 pKa = 4.26DD327 pKa = 4.99VIEE330 pKa = 4.34AWDD333 pKa = 3.46AAGIFSMWANGNSGPACNTSGAPGSRR359 pKa = 11.84IITYY363 pKa = 10.34SVGNYY368 pKa = 9.23NSSHH372 pKa = 6.8AISNTSGKK380 pKa = 10.58GAGQDD385 pKa = 4.11GEE387 pKa = 4.4IKK389 pKa = 10.53PNIAAPGSAIRR400 pKa = 11.84SSVPGNGYY408 pKa = 10.96ANYY411 pKa = 9.79SGTSMASPHH420 pKa = 6.29AAGAVALLWSAAPSLVGDD438 pKa = 3.92IEE440 pKa = 4.42GTRR443 pKa = 11.84ALLDD447 pKa = 3.21GTAIDD452 pKa = 4.75TEE454 pKa = 4.17NLTCGGTADD463 pKa = 4.16DD464 pKa = 4.36NNVFGEE470 pKa = 4.34GRR472 pKa = 11.84LDD474 pKa = 3.45ALALIEE480 pKa = 4.46AAPTGDD486 pKa = 3.27TGEE489 pKa = 4.41LNGTVTDD496 pKa = 4.63ADD498 pKa = 3.7TGDD501 pKa = 4.69AISGANVVIVGEE513 pKa = 4.25AEE515 pKa = 3.98RR516 pKa = 11.84TTVTDD521 pKa = 3.55AEE523 pKa = 4.59GTYY526 pKa = 10.85SSLLVAGDD534 pKa = 3.63YY535 pKa = 11.21DD536 pKa = 3.97VTVSKK541 pKa = 10.55FGYY544 pKa = 8.34GTEE547 pKa = 4.39TEE549 pKa = 4.68SVTVPAGGTATLDD562 pKa = 3.49FDD564 pKa = 5.97LEE566 pKa = 4.42AQEE569 pKa = 4.35STAVSGVVTDD579 pKa = 4.68GSGHH583 pKa = 6.18GWPVYY588 pKa = 10.87AKK590 pKa = 10.6VSVQGTDD597 pKa = 2.5VSTYY601 pKa = 9.08TDD603 pKa = 3.97PATGEE608 pKa = 4.08YY609 pKa = 10.02TLALPIGADD618 pKa = 3.27YY619 pKa = 10.12TLEE622 pKa = 4.26VEE624 pKa = 4.34AQYY627 pKa = 10.76PGYY630 pKa = 8.62EE631 pKa = 4.01TATVVVSPQGAPLTGSRR648 pKa = 11.84EE649 pKa = 3.98SAKK652 pKa = 10.18RR653 pKa = 11.84VPQVVQDD660 pKa = 3.34IQVIVDD666 pKa = 3.88ADD668 pKa = 3.77SCVAPGYY675 pKa = 9.31AHH677 pKa = 6.71SSEE680 pKa = 4.22GVVEE684 pKa = 4.59SFDD687 pKa = 3.55GTEE690 pKa = 4.14TPEE693 pKa = 3.86GWTVTDD699 pKa = 4.28DD700 pKa = 3.71AGTGQVWLFDD710 pKa = 4.11DD711 pKa = 3.7SRR713 pKa = 11.84NRR715 pKa = 11.84GNLTGGEE722 pKa = 3.84GGYY725 pKa = 10.83AIVDD729 pKa = 3.5SDD731 pKa = 4.02NYY733 pKa = 11.11GSGGVQDD740 pKa = 4.2TSLVTPVVDD749 pKa = 4.15MSALAEE755 pKa = 3.83PVIGFKK761 pKa = 10.26HH762 pKa = 6.99DD763 pKa = 3.55YY764 pKa = 10.4NAFLGTGEE772 pKa = 4.16SGEE775 pKa = 4.15VDD777 pKa = 3.41VSIDD781 pKa = 4.25GGEE784 pKa = 3.99TWEE787 pKa = 4.23TLATYY792 pKa = 7.64TTDD795 pKa = 2.88SRR797 pKa = 11.84GEE799 pKa = 4.14VVLPMPTAAGEE810 pKa = 4.02SDD812 pKa = 3.75VQVRR816 pKa = 11.84FHH818 pKa = 6.96YY819 pKa = 10.24SASWDD824 pKa = 3.59YY825 pKa = 8.9WWQVDD830 pKa = 4.49DD831 pKa = 4.34VFVGNRR837 pKa = 11.84TCEE840 pKa = 3.63PSIAGGYY847 pKa = 8.72VVGTVASAEE856 pKa = 4.16TGEE859 pKa = 4.68GIVGASVTSVEE870 pKa = 4.34APEE873 pKa = 4.42EE874 pKa = 4.22SATTVATPDD883 pKa = 5.03DD884 pKa = 4.37EE885 pKa = 5.26NLDD888 pKa = 4.13DD889 pKa = 4.03GFYY892 pKa = 10.19WIFSTLTGAHH902 pKa = 6.65DD903 pKa = 4.5FEE905 pKa = 4.74ATARR909 pKa = 11.84GYY911 pKa = 11.12EE912 pKa = 4.27SVTQSVDD919 pKa = 3.21VAEE922 pKa = 5.14DD923 pKa = 3.55DD924 pKa = 4.09AVRR927 pKa = 11.84ADD929 pKa = 3.6FTLGSGLVDD938 pKa = 3.49VSPDD942 pKa = 3.43SITTYY947 pKa = 10.5VQLGDD952 pKa = 3.86SVTDD956 pKa = 3.78KK957 pKa = 10.76LTISNTGTGAAEE969 pKa = 4.08VTLSEE974 pKa = 4.23LAGDD978 pKa = 4.11FEE980 pKa = 4.9ILRR983 pKa = 11.84ADD985 pKa = 3.57GTKK988 pKa = 10.48EE989 pKa = 4.28LMSEE993 pKa = 3.81MHH995 pKa = 5.86KK996 pKa = 9.77TEE998 pKa = 4.23GAPLQEE1004 pKa = 4.53IEE1006 pKa = 4.83AEE1008 pKa = 4.3VSFAQHH1014 pKa = 6.31ASGLAAQAEE1023 pKa = 4.29PAARR1027 pKa = 11.84GVAEE1031 pKa = 4.94APWTDD1036 pKa = 3.65LPQLGSVNMDD1046 pKa = 2.61GRR1048 pKa = 11.84AVNLDD1053 pKa = 3.49GTWYY1057 pKa = 10.68LITGGSGSASYY1068 pKa = 11.59ADD1070 pKa = 3.68VLRR1073 pKa = 11.84YY1074 pKa = 9.63DD1075 pKa = 4.34AEE1077 pKa = 4.45DD1078 pKa = 3.7MSWTAVAPLPSARR1091 pKa = 11.84NAVTAGVVNGQIVVSGGWVAAGTTAEE1117 pKa = 4.15TFVYY1121 pKa = 10.68DD1122 pKa = 4.43EE1123 pKa = 5.42GADD1126 pKa = 3.32SWTAVADD1133 pKa = 3.72NPVAVSAAGQAVADD1147 pKa = 4.15GKK1149 pKa = 10.74LYY1151 pKa = 11.03SVGGCSSADD1160 pKa = 3.53CTPMSNAVTAYY1171 pKa = 10.61DD1172 pKa = 4.24PGTDD1176 pKa = 3.05TWEE1179 pKa = 4.04TLANYY1184 pKa = 7.54PAPVAFASCGGINGEE1199 pKa = 4.61VYY1201 pKa = 9.44CTGGNGGAAGTAASYY1216 pKa = 11.22VYY1218 pKa = 10.53TPDD1221 pKa = 3.21TDD1223 pKa = 3.04TWTAIADD1230 pKa = 3.79APIDD1234 pKa = 3.62TWASQYY1240 pKa = 11.21AAANGMLIVNGGVQGGTITNATFAYY1265 pKa = 10.29DD1266 pKa = 3.62PSTDD1270 pKa = 3.12EE1271 pKa = 5.12WVDD1274 pKa = 3.84LPNSNTAVYY1283 pKa = 9.86RR1284 pKa = 11.84GAAACGFGKK1293 pKa = 10.67FGGSSGNFNATANTEE1308 pKa = 4.12YY1309 pKa = 11.08LPGFDD1314 pKa = 4.45DD1315 pKa = 5.08CGAAGADD1322 pKa = 3.99VEE1324 pKa = 4.49WLSLSATEE1332 pKa = 4.03LTIPAGEE1339 pKa = 4.38SVTVDD1344 pKa = 2.97VTTDD1348 pKa = 3.22GNVAQPGVYY1357 pKa = 8.65TAGIKK1362 pKa = 10.33VSANTPQSFDD1372 pKa = 4.45PIPVTMHH1379 pKa = 5.99VSPPRR1384 pKa = 11.84PWGKK1388 pKa = 10.83LMGNVSGVDD1397 pKa = 3.78CDD1399 pKa = 3.94TVTAPIAGATVDD1411 pKa = 3.43ITPTRR1416 pKa = 11.84TDD1418 pKa = 2.8HH1419 pKa = 6.63PGWVLSTDD1427 pKa = 3.23ADD1429 pKa = 3.3GDD1431 pKa = 3.93YY1432 pKa = 10.28ATWVNTQHH1440 pKa = 5.9TRR1442 pKa = 11.84PGGVQIIAAKK1452 pKa = 10.48DD1453 pKa = 3.2GWRR1456 pKa = 11.84PKK1458 pKa = 10.6AATADD1463 pKa = 3.79PARR1466 pKa = 11.84GVVRR1470 pKa = 11.84TVNFTLAKK1478 pKa = 9.76IGCC1481 pKa = 4.1
MM1 pKa = 7.5AVHH4 pKa = 6.16GHH6 pKa = 5.92RR7 pKa = 11.84ARR9 pKa = 11.84RR10 pKa = 11.84WLAAVAGASLVASGFGLAGASAQPPQPMAEE40 pKa = 3.86VGATDD45 pKa = 4.83KK46 pKa = 10.92IDD48 pKa = 3.8EE49 pKa = 4.35EE50 pKa = 4.67VSAQIEE56 pKa = 4.4EE57 pKa = 4.23KK58 pKa = 10.64GAADD62 pKa = 3.71FWVRR66 pKa = 11.84FDD68 pKa = 4.24GRR70 pKa = 11.84PDD72 pKa = 3.1MSQFSSISDD81 pKa = 3.05WDD83 pKa = 3.51ARR85 pKa = 11.84GQAVYY90 pKa = 10.66DD91 pKa = 4.13ALTEE95 pKa = 4.16SADD98 pKa = 4.32ASQQDD103 pKa = 3.16IRR105 pKa = 11.84EE106 pKa = 4.04QLDD109 pKa = 3.47ADD111 pKa = 3.95GVDD114 pKa = 4.4YY115 pKa = 11.03QAFWATNAIRR125 pKa = 11.84ITSGDD130 pKa = 3.3QSLVSTLAADD140 pKa = 3.62SGVEE144 pKa = 4.32GIYY147 pKa = 8.5PTTTYY152 pKa = 8.86EE153 pKa = 4.0APEE156 pKa = 3.97LVEE159 pKa = 4.92ADD161 pKa = 4.03RR162 pKa = 11.84EE163 pKa = 4.23MSPSAVEE170 pKa = 3.77WGVADD175 pKa = 4.75INADD179 pKa = 4.1DD180 pKa = 3.43VWSTYY185 pKa = 9.22GVHH188 pKa = 6.84GEE190 pKa = 4.35GIVVGSIDD198 pKa = 4.3TGVQWDD204 pKa = 4.19HH205 pKa = 6.93PALVDD210 pKa = 4.72AYY212 pKa = 10.43RR213 pKa = 11.84GNNGDD218 pKa = 3.42GTFTHH223 pKa = 7.61DD224 pKa = 3.92YY225 pKa = 9.95NWYY228 pKa = 9.31DD229 pKa = 3.43AAGTSPEE236 pKa = 4.22VPADD240 pKa = 3.81GNGHH244 pKa = 4.99GTHH247 pKa = 5.45VTGTMVGDD255 pKa = 4.21DD256 pKa = 3.53GGEE259 pKa = 4.11NQVGVAPGATWISANGCCPSDD280 pKa = 3.48QALIDD285 pKa = 3.6SGQWMLAPTKK295 pKa = 10.68LDD297 pKa = 3.24GTSPDD302 pKa = 3.36TTKK305 pKa = 10.66RR306 pKa = 11.84PHH308 pKa = 7.13IINNSWGTTLPSNDD322 pKa = 3.57PFMEE326 pKa = 4.26DD327 pKa = 4.99VIEE330 pKa = 4.34AWDD333 pKa = 3.46AAGIFSMWANGNSGPACNTSGAPGSRR359 pKa = 11.84IITYY363 pKa = 10.34SVGNYY368 pKa = 9.23NSSHH372 pKa = 6.8AISNTSGKK380 pKa = 10.58GAGQDD385 pKa = 4.11GEE387 pKa = 4.4IKK389 pKa = 10.53PNIAAPGSAIRR400 pKa = 11.84SSVPGNGYY408 pKa = 10.96ANYY411 pKa = 9.79SGTSMASPHH420 pKa = 6.29AAGAVALLWSAAPSLVGDD438 pKa = 3.92IEE440 pKa = 4.42GTRR443 pKa = 11.84ALLDD447 pKa = 3.21GTAIDD452 pKa = 4.75TEE454 pKa = 4.17NLTCGGTADD463 pKa = 4.16DD464 pKa = 4.36NNVFGEE470 pKa = 4.34GRR472 pKa = 11.84LDD474 pKa = 3.45ALALIEE480 pKa = 4.46AAPTGDD486 pKa = 3.27TGEE489 pKa = 4.41LNGTVTDD496 pKa = 4.63ADD498 pKa = 3.7TGDD501 pKa = 4.69AISGANVVIVGEE513 pKa = 4.25AEE515 pKa = 3.98RR516 pKa = 11.84TTVTDD521 pKa = 3.55AEE523 pKa = 4.59GTYY526 pKa = 10.85SSLLVAGDD534 pKa = 3.63YY535 pKa = 11.21DD536 pKa = 3.97VTVSKK541 pKa = 10.55FGYY544 pKa = 8.34GTEE547 pKa = 4.39TEE549 pKa = 4.68SVTVPAGGTATLDD562 pKa = 3.49FDD564 pKa = 5.97LEE566 pKa = 4.42AQEE569 pKa = 4.35STAVSGVVTDD579 pKa = 4.68GSGHH583 pKa = 6.18GWPVYY588 pKa = 10.87AKK590 pKa = 10.6VSVQGTDD597 pKa = 2.5VSTYY601 pKa = 9.08TDD603 pKa = 3.97PATGEE608 pKa = 4.08YY609 pKa = 10.02TLALPIGADD618 pKa = 3.27YY619 pKa = 10.12TLEE622 pKa = 4.26VEE624 pKa = 4.34AQYY627 pKa = 10.76PGYY630 pKa = 8.62EE631 pKa = 4.01TATVVVSPQGAPLTGSRR648 pKa = 11.84EE649 pKa = 3.98SAKK652 pKa = 10.18RR653 pKa = 11.84VPQVVQDD660 pKa = 3.34IQVIVDD666 pKa = 3.88ADD668 pKa = 3.77SCVAPGYY675 pKa = 9.31AHH677 pKa = 6.71SSEE680 pKa = 4.22GVVEE684 pKa = 4.59SFDD687 pKa = 3.55GTEE690 pKa = 4.14TPEE693 pKa = 3.86GWTVTDD699 pKa = 4.28DD700 pKa = 3.71AGTGQVWLFDD710 pKa = 4.11DD711 pKa = 3.7SRR713 pKa = 11.84NRR715 pKa = 11.84GNLTGGEE722 pKa = 3.84GGYY725 pKa = 10.83AIVDD729 pKa = 3.5SDD731 pKa = 4.02NYY733 pKa = 11.11GSGGVQDD740 pKa = 4.2TSLVTPVVDD749 pKa = 4.15MSALAEE755 pKa = 3.83PVIGFKK761 pKa = 10.26HH762 pKa = 6.99DD763 pKa = 3.55YY764 pKa = 10.4NAFLGTGEE772 pKa = 4.16SGEE775 pKa = 4.15VDD777 pKa = 3.41VSIDD781 pKa = 4.25GGEE784 pKa = 3.99TWEE787 pKa = 4.23TLATYY792 pKa = 7.64TTDD795 pKa = 2.88SRR797 pKa = 11.84GEE799 pKa = 4.14VVLPMPTAAGEE810 pKa = 4.02SDD812 pKa = 3.75VQVRR816 pKa = 11.84FHH818 pKa = 6.96YY819 pKa = 10.24SASWDD824 pKa = 3.59YY825 pKa = 8.9WWQVDD830 pKa = 4.49DD831 pKa = 4.34VFVGNRR837 pKa = 11.84TCEE840 pKa = 3.63PSIAGGYY847 pKa = 8.72VVGTVASAEE856 pKa = 4.16TGEE859 pKa = 4.68GIVGASVTSVEE870 pKa = 4.34APEE873 pKa = 4.42EE874 pKa = 4.22SATTVATPDD883 pKa = 5.03DD884 pKa = 4.37EE885 pKa = 5.26NLDD888 pKa = 4.13DD889 pKa = 4.03GFYY892 pKa = 10.19WIFSTLTGAHH902 pKa = 6.65DD903 pKa = 4.5FEE905 pKa = 4.74ATARR909 pKa = 11.84GYY911 pKa = 11.12EE912 pKa = 4.27SVTQSVDD919 pKa = 3.21VAEE922 pKa = 5.14DD923 pKa = 3.55DD924 pKa = 4.09AVRR927 pKa = 11.84ADD929 pKa = 3.6FTLGSGLVDD938 pKa = 3.49VSPDD942 pKa = 3.43SITTYY947 pKa = 10.5VQLGDD952 pKa = 3.86SVTDD956 pKa = 3.78KK957 pKa = 10.76LTISNTGTGAAEE969 pKa = 4.08VTLSEE974 pKa = 4.23LAGDD978 pKa = 4.11FEE980 pKa = 4.9ILRR983 pKa = 11.84ADD985 pKa = 3.57GTKK988 pKa = 10.48EE989 pKa = 4.28LMSEE993 pKa = 3.81MHH995 pKa = 5.86KK996 pKa = 9.77TEE998 pKa = 4.23GAPLQEE1004 pKa = 4.53IEE1006 pKa = 4.83AEE1008 pKa = 4.3VSFAQHH1014 pKa = 6.31ASGLAAQAEE1023 pKa = 4.29PAARR1027 pKa = 11.84GVAEE1031 pKa = 4.94APWTDD1036 pKa = 3.65LPQLGSVNMDD1046 pKa = 2.61GRR1048 pKa = 11.84AVNLDD1053 pKa = 3.49GTWYY1057 pKa = 10.68LITGGSGSASYY1068 pKa = 11.59ADD1070 pKa = 3.68VLRR1073 pKa = 11.84YY1074 pKa = 9.63DD1075 pKa = 4.34AEE1077 pKa = 4.45DD1078 pKa = 3.7MSWTAVAPLPSARR1091 pKa = 11.84NAVTAGVVNGQIVVSGGWVAAGTTAEE1117 pKa = 4.15TFVYY1121 pKa = 10.68DD1122 pKa = 4.43EE1123 pKa = 5.42GADD1126 pKa = 3.32SWTAVADD1133 pKa = 3.72NPVAVSAAGQAVADD1147 pKa = 4.15GKK1149 pKa = 10.74LYY1151 pKa = 11.03SVGGCSSADD1160 pKa = 3.53CTPMSNAVTAYY1171 pKa = 10.61DD1172 pKa = 4.24PGTDD1176 pKa = 3.05TWEE1179 pKa = 4.04TLANYY1184 pKa = 7.54PAPVAFASCGGINGEE1199 pKa = 4.61VYY1201 pKa = 9.44CTGGNGGAAGTAASYY1216 pKa = 11.22VYY1218 pKa = 10.53TPDD1221 pKa = 3.21TDD1223 pKa = 3.04TWTAIADD1230 pKa = 3.79APIDD1234 pKa = 3.62TWASQYY1240 pKa = 11.21AAANGMLIVNGGVQGGTITNATFAYY1265 pKa = 10.29DD1266 pKa = 3.62PSTDD1270 pKa = 3.12EE1271 pKa = 5.12WVDD1274 pKa = 3.84LPNSNTAVYY1283 pKa = 9.86RR1284 pKa = 11.84GAAACGFGKK1293 pKa = 10.67FGGSSGNFNATANTEE1308 pKa = 4.12YY1309 pKa = 11.08LPGFDD1314 pKa = 4.45DD1315 pKa = 5.08CGAAGADD1322 pKa = 3.99VEE1324 pKa = 4.49WLSLSATEE1332 pKa = 4.03LTIPAGEE1339 pKa = 4.38SVTVDD1344 pKa = 2.97VTTDD1348 pKa = 3.22GNVAQPGVYY1357 pKa = 8.65TAGIKK1362 pKa = 10.33VSANTPQSFDD1372 pKa = 4.45PIPVTMHH1379 pKa = 5.99VSPPRR1384 pKa = 11.84PWGKK1388 pKa = 10.83LMGNVSGVDD1397 pKa = 3.78CDD1399 pKa = 3.94TVTAPIAGATVDD1411 pKa = 3.43ITPTRR1416 pKa = 11.84TDD1418 pKa = 2.8HH1419 pKa = 6.63PGWVLSTDD1427 pKa = 3.23ADD1429 pKa = 3.3GDD1431 pKa = 3.93YY1432 pKa = 10.28ATWVNTQHH1440 pKa = 5.9TRR1442 pKa = 11.84PGGVQIIAAKK1452 pKa = 10.48DD1453 pKa = 3.2GWRR1456 pKa = 11.84PKK1458 pKa = 10.6AATADD1463 pKa = 3.79PARR1466 pKa = 11.84GVVRR1470 pKa = 11.84TVNFTLAKK1478 pKa = 9.76IGCC1481 pKa = 4.1
Molecular weight: 152.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A516G6M8|A0A516G6M8_9MICO Formyltetrahydrofolate deformylase OS=Ornithinimicrobium sp. H23M54 OX=2594265 GN=purU PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SRR15 pKa = 11.84VHH17 pKa = 6.33GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILASRR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.66GRR40 pKa = 11.84TKK42 pKa = 10.86LSAA45 pKa = 3.61
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SRR15 pKa = 11.84VHH17 pKa = 6.33GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILASRR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.66GRR40 pKa = 11.84TKK42 pKa = 10.86LSAA45 pKa = 3.61
Molecular weight: 5.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1205281 |
32 |
3158 |
332.8 |
35.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.637 ± 0.048 | 0.667 ± 0.011 |
6.3 ± 0.039 | 6.06 ± 0.038 |
2.653 ± 0.024 | 9.27 ± 0.039 |
2.299 ± 0.021 | 3.707 ± 0.031 |
1.662 ± 0.026 | 10.395 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.887 ± 0.018 | 1.741 ± 0.021 |
5.744 ± 0.031 | 3.3 ± 0.019 |
7.277 ± 0.049 | 5.458 ± 0.033 |
6.414 ± 0.038 | 9.06 ± 0.036 |
1.589 ± 0.015 | 1.88 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
