
Apple chlorotic leaf spot virus (isolate apple) (ACLSV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Trivirinae; Trichovirus; Apple chlorotic leaf spot virus
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
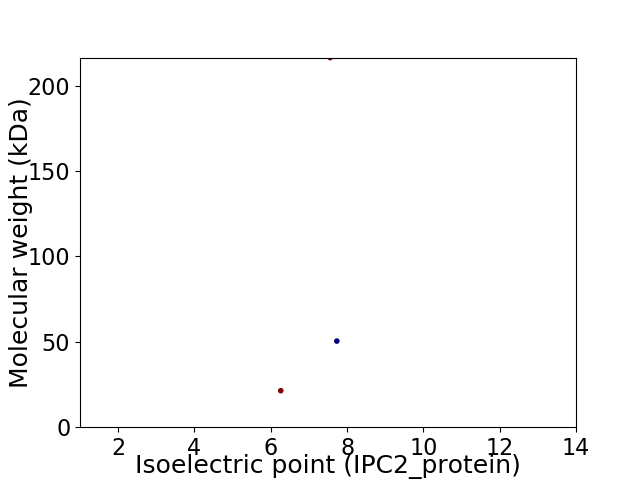
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
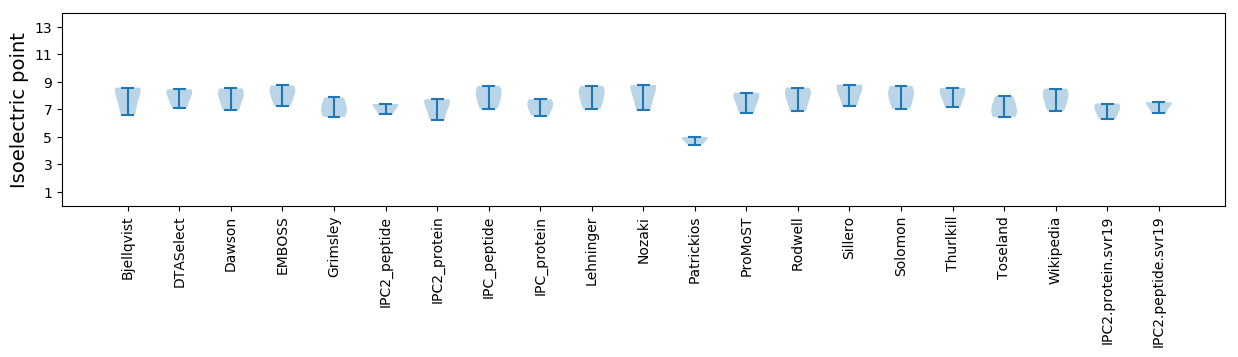
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P54891|RDRP_ACLSA RNA-directed RNA polymerase OS=Apple chlorotic leaf spot virus (isolate apple) OX=73472 PE=4 SV=1
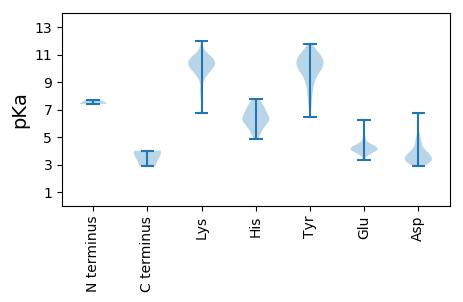
MM1 pKa = 7.42AAVLNLQLKK10 pKa = 9.93VDD12 pKa = 4.05ADD14 pKa = 4.15LKK16 pKa = 11.28AFLAAEE22 pKa = 4.21GRR24 pKa = 11.84PLHH27 pKa = 6.35GKK29 pKa = 7.75TGAILEE35 pKa = 4.3QTLEE39 pKa = 4.37AIFANIAIQGTSEE52 pKa = 3.75QTEE55 pKa = 4.1FLDD58 pKa = 3.45VLVEE62 pKa = 4.23VKK64 pKa = 10.81SMEE67 pKa = 4.09DD68 pKa = 3.16QKK70 pKa = 11.74VVGSFNLKK78 pKa = 9.94EE79 pKa = 3.97VVGLIKK85 pKa = 10.26IFRR88 pKa = 11.84TTSSDD93 pKa = 3.56PNISSMTFRR102 pKa = 11.84QVCEE106 pKa = 3.82AFAPEE111 pKa = 4.08ARR113 pKa = 11.84NGLVKK118 pKa = 10.76LKK120 pKa = 10.82YY121 pKa = 10.13KK122 pKa = 10.81GVFTNLFSTTPEE134 pKa = 3.9VGGKK138 pKa = 9.94YY139 pKa = 9.57PEE141 pKa = 5.03LMFDD145 pKa = 4.06FNKK148 pKa = 9.68GLNMFIMNKK157 pKa = 9.41AQQKK161 pKa = 10.48VITNMNRR168 pKa = 11.84RR169 pKa = 11.84LLQTEE174 pKa = 4.41FAKK177 pKa = 10.83SEE179 pKa = 4.15NEE181 pKa = 3.7AKK183 pKa = 9.72MSSVTTDD190 pKa = 3.07LCVV193 pKa = 2.87
MM1 pKa = 7.42AAVLNLQLKK10 pKa = 9.93VDD12 pKa = 4.05ADD14 pKa = 4.15LKK16 pKa = 11.28AFLAAEE22 pKa = 4.21GRR24 pKa = 11.84PLHH27 pKa = 6.35GKK29 pKa = 7.75TGAILEE35 pKa = 4.3QTLEE39 pKa = 4.37AIFANIAIQGTSEE52 pKa = 3.75QTEE55 pKa = 4.1FLDD58 pKa = 3.45VLVEE62 pKa = 4.23VKK64 pKa = 10.81SMEE67 pKa = 4.09DD68 pKa = 3.16QKK70 pKa = 11.74VVGSFNLKK78 pKa = 9.94EE79 pKa = 3.97VVGLIKK85 pKa = 10.26IFRR88 pKa = 11.84TTSSDD93 pKa = 3.56PNISSMTFRR102 pKa = 11.84QVCEE106 pKa = 3.82AFAPEE111 pKa = 4.08ARR113 pKa = 11.84NGLVKK118 pKa = 10.76LKK120 pKa = 10.82YY121 pKa = 10.13KK122 pKa = 10.81GVFTNLFSTTPEE134 pKa = 3.9VGGKK138 pKa = 9.94YY139 pKa = 9.57PEE141 pKa = 5.03LMFDD145 pKa = 4.06FNKK148 pKa = 9.68GLNMFIMNKK157 pKa = 9.41AQQKK161 pKa = 10.48VITNMNRR168 pKa = 11.84RR169 pKa = 11.84LLQTEE174 pKa = 4.41FAKK177 pKa = 10.83SEE179 pKa = 4.15NEE181 pKa = 3.7AKK183 pKa = 9.72MSSVTTDD190 pKa = 3.07LCVV193 pKa = 2.87
Molecular weight: 21.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P54892|MP_ACLSA Putative movement protein OS=Apple chlorotic leaf spot virus (isolate apple) OX=73472 PE=3 SV=1
MM1 pKa = 7.72MIRR4 pKa = 11.84GHH6 pKa = 5.93KK7 pKa = 10.14LKK9 pKa = 10.23IAEE12 pKa = 4.25GDD14 pKa = 3.48IPIAGVKK21 pKa = 9.81SSRR24 pKa = 11.84IYY26 pKa = 11.03SDD28 pKa = 3.02VSPFRR33 pKa = 11.84KK34 pKa = 10.12ASDD37 pKa = 4.92LMIHH41 pKa = 5.83WNEE44 pKa = 3.84FVFKK48 pKa = 10.88VMPEE52 pKa = 4.42DD53 pKa = 3.29IAGDD57 pKa = 4.15GFRR60 pKa = 11.84LASIPVIPSSEE71 pKa = 4.05VQAVLRR77 pKa = 11.84KK78 pKa = 9.69RR79 pKa = 11.84EE80 pKa = 4.1STNYY84 pKa = 7.76VHH86 pKa = 7.3WGALSISIDD95 pKa = 3.16ALFRR99 pKa = 11.84KK100 pKa = 9.49NAGVSGWCYY109 pKa = 10.45VYY111 pKa = 11.0DD112 pKa = 3.98NRR114 pKa = 11.84WEE116 pKa = 4.18TFEE119 pKa = 4.38QAMLQKK125 pKa = 10.47FRR127 pKa = 11.84FNLDD131 pKa = 3.1SGSATLVTSPNFPVSLDD148 pKa = 3.76DD149 pKa = 4.79PGLSNSISVAVMFEE163 pKa = 3.79NLNFKK168 pKa = 10.66LEE170 pKa = 4.24SYY172 pKa = 9.46PISVRR177 pKa = 11.84VGNMCRR183 pKa = 11.84FFDD186 pKa = 4.65SFLSCVKK193 pKa = 10.78NKK195 pKa = 9.87VDD197 pKa = 5.0SNFLLEE203 pKa = 4.68AANADD208 pKa = 3.96PLGAGAFGFEE218 pKa = 4.23QDD220 pKa = 4.16DD221 pKa = 3.85QVSEE225 pKa = 4.25LFNYY229 pKa = 8.89IQTVPTQAIKK239 pKa = 10.65SRR241 pKa = 11.84EE242 pKa = 4.09HH243 pKa = 5.44EE244 pKa = 4.27VPRR247 pKa = 11.84GLFGMMGKK255 pKa = 9.96KK256 pKa = 9.39KK257 pKa = 10.4VKK259 pKa = 10.24SFEE262 pKa = 4.07FASGSRR268 pKa = 11.84NLGRR272 pKa = 11.84RR273 pKa = 11.84KK274 pKa = 8.91PQRR277 pKa = 11.84GRR279 pKa = 11.84PLEE282 pKa = 4.14RR283 pKa = 11.84SASLRR288 pKa = 11.84VAPGFRR294 pKa = 11.84SQDD297 pKa = 2.94EE298 pKa = 4.4RR299 pKa = 11.84VEE301 pKa = 4.13HH302 pKa = 6.25QGLSTDD308 pKa = 3.31SDD310 pKa = 3.95FEE312 pKa = 4.18NFLRR316 pKa = 11.84KK317 pKa = 9.94GKK319 pKa = 10.4GKK321 pKa = 10.5AGTEE325 pKa = 4.55SITSEE330 pKa = 3.89GSSFDD335 pKa = 3.91NISARR340 pKa = 11.84EE341 pKa = 3.87FQFARR346 pKa = 11.84QDD348 pKa = 3.42QKK350 pKa = 12.0AKK352 pKa = 10.64DD353 pKa = 3.82GGSAEE358 pKa = 4.32PPIKK362 pKa = 10.14SGRR365 pKa = 11.84RR366 pKa = 11.84SEE368 pKa = 4.39SVPGRR373 pKa = 11.84RR374 pKa = 11.84RR375 pKa = 11.84QTPSWKK381 pKa = 10.16DD382 pKa = 3.22RR383 pKa = 11.84GNSGTDD389 pKa = 2.9TGGHH393 pKa = 5.69LRR395 pKa = 11.84EE396 pKa = 4.7HH397 pKa = 7.27SDD399 pKa = 3.41TGDD402 pKa = 3.25LGTNRR407 pKa = 11.84VPGRR411 pKa = 11.84AGGGEE416 pKa = 3.57IHH418 pKa = 6.72GGSEE422 pKa = 4.15GGGVIQSEE430 pKa = 4.44GGSRR434 pKa = 11.84FDD436 pKa = 5.53QNIQDD441 pKa = 4.16YY442 pKa = 11.0IFGPEE447 pKa = 4.13YY448 pKa = 9.96KK449 pKa = 10.09QHH451 pKa = 7.13DD452 pKa = 4.65LPPSVV457 pKa = 3.85
MM1 pKa = 7.72MIRR4 pKa = 11.84GHH6 pKa = 5.93KK7 pKa = 10.14LKK9 pKa = 10.23IAEE12 pKa = 4.25GDD14 pKa = 3.48IPIAGVKK21 pKa = 9.81SSRR24 pKa = 11.84IYY26 pKa = 11.03SDD28 pKa = 3.02VSPFRR33 pKa = 11.84KK34 pKa = 10.12ASDD37 pKa = 4.92LMIHH41 pKa = 5.83WNEE44 pKa = 3.84FVFKK48 pKa = 10.88VMPEE52 pKa = 4.42DD53 pKa = 3.29IAGDD57 pKa = 4.15GFRR60 pKa = 11.84LASIPVIPSSEE71 pKa = 4.05VQAVLRR77 pKa = 11.84KK78 pKa = 9.69RR79 pKa = 11.84EE80 pKa = 4.1STNYY84 pKa = 7.76VHH86 pKa = 7.3WGALSISIDD95 pKa = 3.16ALFRR99 pKa = 11.84KK100 pKa = 9.49NAGVSGWCYY109 pKa = 10.45VYY111 pKa = 11.0DD112 pKa = 3.98NRR114 pKa = 11.84WEE116 pKa = 4.18TFEE119 pKa = 4.38QAMLQKK125 pKa = 10.47FRR127 pKa = 11.84FNLDD131 pKa = 3.1SGSATLVTSPNFPVSLDD148 pKa = 3.76DD149 pKa = 4.79PGLSNSISVAVMFEE163 pKa = 3.79NLNFKK168 pKa = 10.66LEE170 pKa = 4.24SYY172 pKa = 9.46PISVRR177 pKa = 11.84VGNMCRR183 pKa = 11.84FFDD186 pKa = 4.65SFLSCVKK193 pKa = 10.78NKK195 pKa = 9.87VDD197 pKa = 5.0SNFLLEE203 pKa = 4.68AANADD208 pKa = 3.96PLGAGAFGFEE218 pKa = 4.23QDD220 pKa = 4.16DD221 pKa = 3.85QVSEE225 pKa = 4.25LFNYY229 pKa = 8.89IQTVPTQAIKK239 pKa = 10.65SRR241 pKa = 11.84EE242 pKa = 4.09HH243 pKa = 5.44EE244 pKa = 4.27VPRR247 pKa = 11.84GLFGMMGKK255 pKa = 9.96KK256 pKa = 9.39KK257 pKa = 10.4VKK259 pKa = 10.24SFEE262 pKa = 4.07FASGSRR268 pKa = 11.84NLGRR272 pKa = 11.84RR273 pKa = 11.84KK274 pKa = 8.91PQRR277 pKa = 11.84GRR279 pKa = 11.84PLEE282 pKa = 4.14RR283 pKa = 11.84SASLRR288 pKa = 11.84VAPGFRR294 pKa = 11.84SQDD297 pKa = 2.94EE298 pKa = 4.4RR299 pKa = 11.84VEE301 pKa = 4.13HH302 pKa = 6.25QGLSTDD308 pKa = 3.31SDD310 pKa = 3.95FEE312 pKa = 4.18NFLRR316 pKa = 11.84KK317 pKa = 9.94GKK319 pKa = 10.4GKK321 pKa = 10.5AGTEE325 pKa = 4.55SITSEE330 pKa = 3.89GSSFDD335 pKa = 3.91NISARR340 pKa = 11.84EE341 pKa = 3.87FQFARR346 pKa = 11.84QDD348 pKa = 3.42QKK350 pKa = 12.0AKK352 pKa = 10.64DD353 pKa = 3.82GGSAEE358 pKa = 4.32PPIKK362 pKa = 10.14SGRR365 pKa = 11.84RR366 pKa = 11.84SEE368 pKa = 4.39SVPGRR373 pKa = 11.84RR374 pKa = 11.84RR375 pKa = 11.84QTPSWKK381 pKa = 10.16DD382 pKa = 3.22RR383 pKa = 11.84GNSGTDD389 pKa = 2.9TGGHH393 pKa = 5.69LRR395 pKa = 11.84EE396 pKa = 4.7HH397 pKa = 7.27SDD399 pKa = 3.41TGDD402 pKa = 3.25LGTNRR407 pKa = 11.84VPGRR411 pKa = 11.84AGGGEE416 pKa = 3.57IHH418 pKa = 6.72GGSEE422 pKa = 4.15GGGVIQSEE430 pKa = 4.44GGSRR434 pKa = 11.84FDD436 pKa = 5.53QNIQDD441 pKa = 4.16YY442 pKa = 11.0IFGPEE447 pKa = 4.13YY448 pKa = 9.96KK449 pKa = 10.09QHH451 pKa = 7.13DD452 pKa = 4.65LPPSVV457 pKa = 3.85
Molecular weight: 50.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2535 |
193 |
1885 |
845.0 |
96.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.207 ± 0.727 | 2.091 ± 0.655 |
5.641 ± 0.314 | 7.495 ± 0.407 |
5.838 ± 0.306 | 6.943 ± 1.234 |
2.367 ± 0.43 | 5.957 ± 0.583 |
7.89 ± 0.871 | 8.679 ± 0.921 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.761 ± 0.274 | 4.024 ± 0.478 |
3.708 ± 0.43 | 3.274 ± 0.434 |
6.114 ± 0.557 | 8.481 ± 0.794 |
3.708 ± 0.559 | 5.838 ± 0.507 |
1.065 ± 0.187 | 2.919 ± 0.677 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
