
Human metapneumovirus (strain CAN97-83) (HMPV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Pneumoviridae; Metapneumovirus; Human metapneumovirus
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
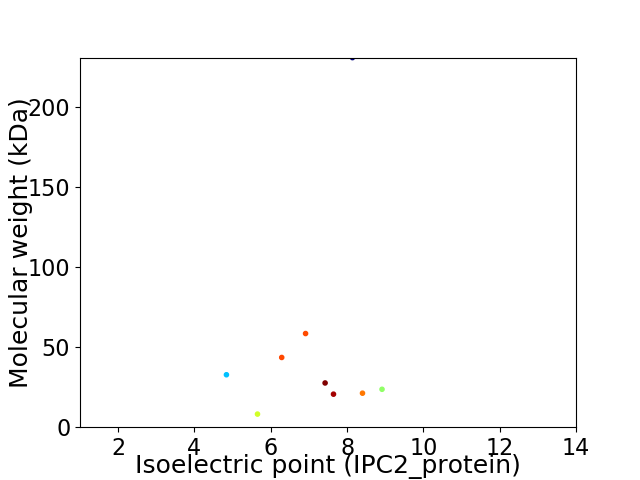
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q8B9Q8|PHOSP_HMPVC Phosphoprotein OS=Human metapneumovirus (strain CAN97-83) OX=694067 GN=P PE=1 SV=1
MM1 pKa = 7.55SFPEE5 pKa = 4.49GKK7 pKa = 10.16DD8 pKa = 2.84ILFMGNEE15 pKa = 3.85AAKK18 pKa = 10.34LAEE21 pKa = 4.55AFQKK25 pKa = 10.38SLRR28 pKa = 11.84KK29 pKa = 9.13PSHH32 pKa = 6.41KK33 pKa = 9.83RR34 pKa = 11.84SQSIIGEE41 pKa = 4.26KK42 pKa = 10.47VNTVSEE48 pKa = 4.29TLEE51 pKa = 4.38LPTISRR57 pKa = 11.84PTKK60 pKa = 8.75PTILSEE66 pKa = 4.38PKK68 pKa = 10.39LAWTDD73 pKa = 2.87KK74 pKa = 10.85GGAIKK79 pKa = 9.81TEE81 pKa = 3.85AKK83 pKa = 8.85QTIKK87 pKa = 10.96VMDD90 pKa = 5.18PIEE93 pKa = 4.26EE94 pKa = 4.2EE95 pKa = 4.03EE96 pKa = 4.18FTEE99 pKa = 4.89KK100 pKa = 10.43RR101 pKa = 11.84VLPSSDD107 pKa = 2.6GKK109 pKa = 9.18TPAEE113 pKa = 4.28KK114 pKa = 10.33KK115 pKa = 10.41LKK117 pKa = 10.37PSTNTKK123 pKa = 9.95KK124 pKa = 10.56KK125 pKa = 10.91VSFTPNEE132 pKa = 3.81PGKK135 pKa = 8.74YY136 pKa = 8.55TKK138 pKa = 10.59LEE140 pKa = 3.97KK141 pKa = 10.45DD142 pKa = 3.58ALDD145 pKa = 3.95LLSDD149 pKa = 3.95NEE151 pKa = 4.16EE152 pKa = 3.86EE153 pKa = 4.39DD154 pKa = 4.05AEE156 pKa = 4.49SSILTFEE163 pKa = 4.34EE164 pKa = 4.82RR165 pKa = 11.84DD166 pKa = 3.58TSSLSIEE173 pKa = 4.18ARR175 pKa = 11.84LEE177 pKa = 4.23SIEE180 pKa = 4.71EE181 pKa = 4.18KK182 pKa = 10.85LSMILGLLRR191 pKa = 11.84TLNIATAGPTAARR204 pKa = 11.84DD205 pKa = 4.16GIRR208 pKa = 11.84DD209 pKa = 3.43AMIGIRR215 pKa = 11.84EE216 pKa = 4.13EE217 pKa = 4.64LIADD221 pKa = 4.3IIKK224 pKa = 9.71EE225 pKa = 4.32AKK227 pKa = 9.86GKK229 pKa = 9.21AAEE232 pKa = 4.1MMEE235 pKa = 4.19EE236 pKa = 4.42EE237 pKa = 4.33MNQRR241 pKa = 11.84TKK243 pKa = 10.55IGNGSVKK250 pKa = 9.54LTEE253 pKa = 4.33KK254 pKa = 10.76AKK256 pKa = 9.89EE257 pKa = 3.84LNKK260 pKa = 10.09IVEE263 pKa = 4.67DD264 pKa = 4.06EE265 pKa = 4.41STSGEE270 pKa = 4.21SEE272 pKa = 4.07EE273 pKa = 4.47EE274 pKa = 4.15EE275 pKa = 4.18EE276 pKa = 5.87LKK278 pKa = 10.09DD279 pKa = 3.58TQEE282 pKa = 4.17NNQEE286 pKa = 3.77DD287 pKa = 5.25DD288 pKa = 3.43IYY290 pKa = 11.19QLIMM294 pKa = 4.27
MM1 pKa = 7.55SFPEE5 pKa = 4.49GKK7 pKa = 10.16DD8 pKa = 2.84ILFMGNEE15 pKa = 3.85AAKK18 pKa = 10.34LAEE21 pKa = 4.55AFQKK25 pKa = 10.38SLRR28 pKa = 11.84KK29 pKa = 9.13PSHH32 pKa = 6.41KK33 pKa = 9.83RR34 pKa = 11.84SQSIIGEE41 pKa = 4.26KK42 pKa = 10.47VNTVSEE48 pKa = 4.29TLEE51 pKa = 4.38LPTISRR57 pKa = 11.84PTKK60 pKa = 8.75PTILSEE66 pKa = 4.38PKK68 pKa = 10.39LAWTDD73 pKa = 2.87KK74 pKa = 10.85GGAIKK79 pKa = 9.81TEE81 pKa = 3.85AKK83 pKa = 8.85QTIKK87 pKa = 10.96VMDD90 pKa = 5.18PIEE93 pKa = 4.26EE94 pKa = 4.2EE95 pKa = 4.03EE96 pKa = 4.18FTEE99 pKa = 4.89KK100 pKa = 10.43RR101 pKa = 11.84VLPSSDD107 pKa = 2.6GKK109 pKa = 9.18TPAEE113 pKa = 4.28KK114 pKa = 10.33KK115 pKa = 10.41LKK117 pKa = 10.37PSTNTKK123 pKa = 9.95KK124 pKa = 10.56KK125 pKa = 10.91VSFTPNEE132 pKa = 3.81PGKK135 pKa = 8.74YY136 pKa = 8.55TKK138 pKa = 10.59LEE140 pKa = 3.97KK141 pKa = 10.45DD142 pKa = 3.58ALDD145 pKa = 3.95LLSDD149 pKa = 3.95NEE151 pKa = 4.16EE152 pKa = 3.86EE153 pKa = 4.39DD154 pKa = 4.05AEE156 pKa = 4.49SSILTFEE163 pKa = 4.34EE164 pKa = 4.82RR165 pKa = 11.84DD166 pKa = 3.58TSSLSIEE173 pKa = 4.18ARR175 pKa = 11.84LEE177 pKa = 4.23SIEE180 pKa = 4.71EE181 pKa = 4.18KK182 pKa = 10.85LSMILGLLRR191 pKa = 11.84TLNIATAGPTAARR204 pKa = 11.84DD205 pKa = 4.16GIRR208 pKa = 11.84DD209 pKa = 3.43AMIGIRR215 pKa = 11.84EE216 pKa = 4.13EE217 pKa = 4.64LIADD221 pKa = 4.3IIKK224 pKa = 9.71EE225 pKa = 4.32AKK227 pKa = 9.86GKK229 pKa = 9.21AAEE232 pKa = 4.1MMEE235 pKa = 4.19EE236 pKa = 4.42EE237 pKa = 4.33MNQRR241 pKa = 11.84TKK243 pKa = 10.55IGNGSVKK250 pKa = 9.54LTEE253 pKa = 4.33KK254 pKa = 10.76AKK256 pKa = 9.89EE257 pKa = 3.84LNKK260 pKa = 10.09IVEE263 pKa = 4.67DD264 pKa = 4.06EE265 pKa = 4.41STSGEE270 pKa = 4.21SEE272 pKa = 4.07EE273 pKa = 4.47EE274 pKa = 4.15EE275 pKa = 4.18EE276 pKa = 5.87LKK278 pKa = 10.09DD279 pKa = 3.58TQEE282 pKa = 4.17NNQEE286 pKa = 3.77DD287 pKa = 5.25DD288 pKa = 3.43IYY290 pKa = 11.19QLIMM294 pKa = 4.27
Molecular weight: 32.75 kDa
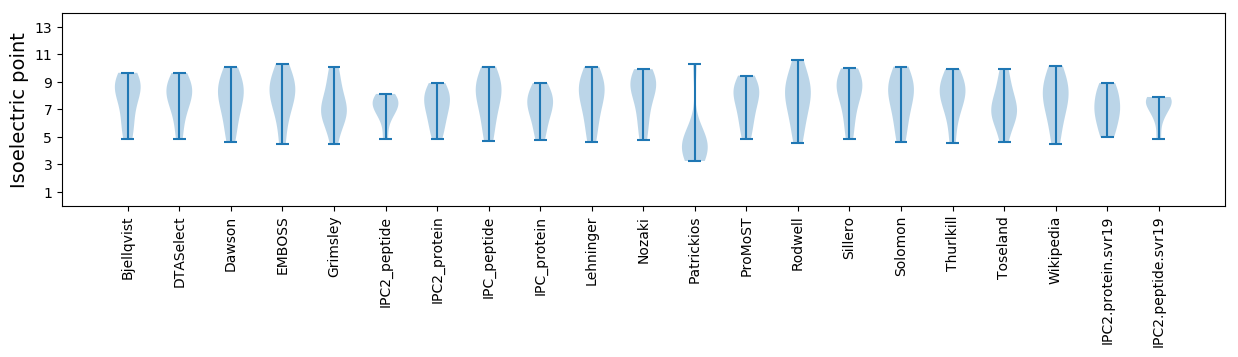
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q6WB95|SH_HMPVC Small hydrophobic protein OS=Human metapneumovirus (strain CAN97-83) OX=694067 GN=SH PE=1 SV=1
MM1 pKa = 7.13EE2 pKa = 4.63VKK4 pKa = 10.43VEE6 pKa = 4.26NIRR9 pKa = 11.84AIDD12 pKa = 3.62MLKK15 pKa = 10.53ARR17 pKa = 11.84VKK19 pKa = 10.9NRR21 pKa = 11.84VARR24 pKa = 11.84SKK26 pKa = 10.7CFKK29 pKa = 10.02NASLILIGITTLSIALNIYY48 pKa = 10.04LIINYY53 pKa = 7.58TIQKK57 pKa = 7.96TSSEE61 pKa = 4.63SEE63 pKa = 4.05HH64 pKa = 6.38HH65 pKa = 6.38TSSPPTEE72 pKa = 4.39SNKK75 pKa = 9.88EE76 pKa = 3.7ASTISTDD83 pKa = 4.23NPDD86 pKa = 3.43INPNSQHH93 pKa = 6.32PTQQSTEE100 pKa = 3.98NPTLNPAASVSPSEE114 pKa = 4.44TEE116 pKa = 3.86PASTPDD122 pKa = 3.14TTNRR126 pKa = 11.84LSSVDD131 pKa = 3.43RR132 pKa = 11.84STAQPSEE139 pKa = 4.29SRR141 pKa = 11.84TKK143 pKa = 10.09TKK145 pKa = 9.36PTVHH149 pKa = 6.0TRR151 pKa = 11.84NNPSTASSTQSPPRR165 pKa = 11.84ATTKK169 pKa = 10.48AIRR172 pKa = 11.84RR173 pKa = 11.84ATTFRR178 pKa = 11.84MSSTGKK184 pKa = 10.35RR185 pKa = 11.84PTTTSVQSDD194 pKa = 3.61SSTTTQNHH202 pKa = 6.17EE203 pKa = 4.37EE204 pKa = 4.16TGSANPQASVSTMQNN219 pKa = 2.73
MM1 pKa = 7.13EE2 pKa = 4.63VKK4 pKa = 10.43VEE6 pKa = 4.26NIRR9 pKa = 11.84AIDD12 pKa = 3.62MLKK15 pKa = 10.53ARR17 pKa = 11.84VKK19 pKa = 10.9NRR21 pKa = 11.84VARR24 pKa = 11.84SKK26 pKa = 10.7CFKK29 pKa = 10.02NASLILIGITTLSIALNIYY48 pKa = 10.04LIINYY53 pKa = 7.58TIQKK57 pKa = 7.96TSSEE61 pKa = 4.63SEE63 pKa = 4.05HH64 pKa = 6.38HH65 pKa = 6.38TSSPPTEE72 pKa = 4.39SNKK75 pKa = 9.88EE76 pKa = 3.7ASTISTDD83 pKa = 4.23NPDD86 pKa = 3.43INPNSQHH93 pKa = 6.32PTQQSTEE100 pKa = 3.98NPTLNPAASVSPSEE114 pKa = 4.44TEE116 pKa = 3.86PASTPDD122 pKa = 3.14TTNRR126 pKa = 11.84LSSVDD131 pKa = 3.43RR132 pKa = 11.84STAQPSEE139 pKa = 4.29SRR141 pKa = 11.84TKK143 pKa = 10.09TKK145 pKa = 9.36PTVHH149 pKa = 6.0TRR151 pKa = 11.84NNPSTASSTQSPPRR165 pKa = 11.84ATTKK169 pKa = 10.48AIRR172 pKa = 11.84RR173 pKa = 11.84ATTFRR178 pKa = 11.84MSSTGKK184 pKa = 10.35RR185 pKa = 11.84PTTTSVQSDD194 pKa = 3.61SSTTTQNHH202 pKa = 6.17EE203 pKa = 4.37EE204 pKa = 4.16TGSANPQASVSTMQNN219 pKa = 2.73
Molecular weight: 23.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4142 |
71 |
2005 |
460.2 |
51.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.674 ± 1.039 | 1.835 ± 0.321 |
4.732 ± 0.292 | 6.663 ± 0.883 |
3.356 ± 0.612 | 4.708 ± 0.55 |
1.787 ± 0.256 | 7.798 ± 0.382 |
7.846 ± 0.606 | 10.092 ± 0.954 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.487 ± 0.277 | 5.529 ± 0.464 |
3.597 ± 0.439 | 3.573 ± 0.295 |
4.491 ± 0.479 | 8.788 ± 0.839 |
6.808 ± 1.096 | 5.963 ± 0.589 |
1.062 ± 0.243 | 3.211 ± 0.395 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
