
Hymenobacter jejuensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Hymenobacteraceae; Hymenobacter
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
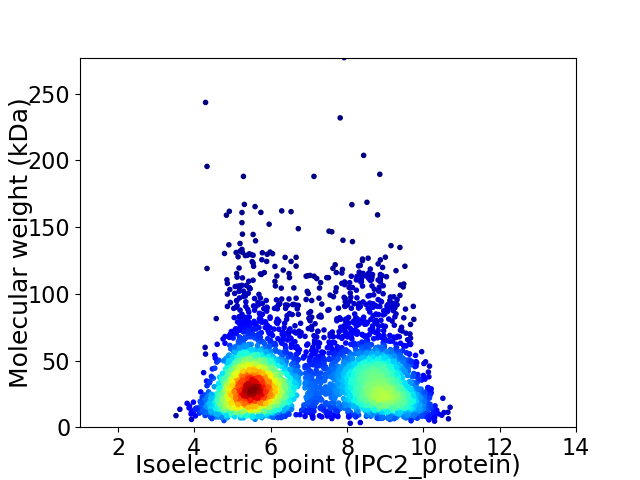
Virtual 2D-PAGE plot for 4100 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
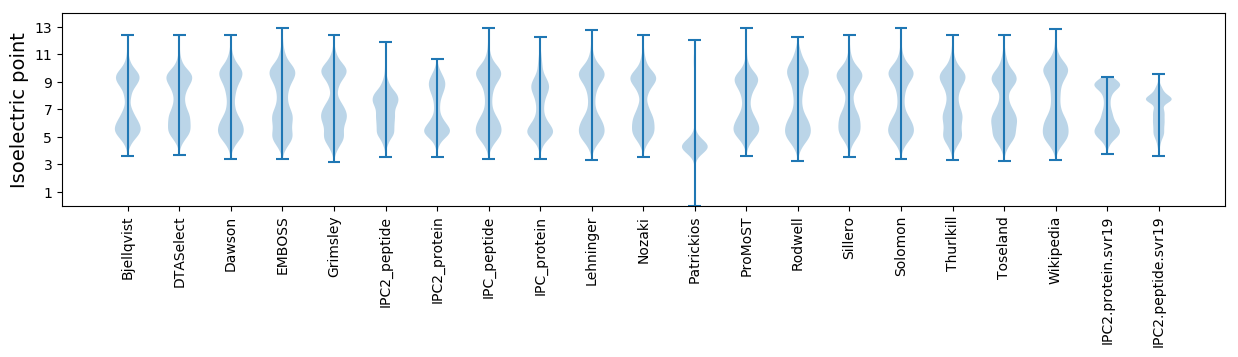
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B8A0S9|A0A5B8A0S9_9BACT GSH-S_ATP domain-containing protein OS=Hymenobacter jejuensis OX=2502781 GN=FHG12_09715 PE=4 SV=1
MM1 pKa = 7.39KK2 pKa = 10.48RR3 pKa = 11.84LIDD6 pKa = 4.37ALADD10 pKa = 3.86DD11 pKa = 4.83ADD13 pKa = 4.09FFVEE17 pKa = 4.74HH18 pKa = 5.67VQITAIVFDD27 pKa = 3.77NTNDD31 pKa = 3.3VTVWATTYY39 pKa = 10.58FDD41 pKa = 3.89EE42 pKa = 4.67EE43 pKa = 4.34LHH45 pKa = 6.19FFHH48 pKa = 7.36LGLQFQYY55 pKa = 11.47LDD57 pKa = 3.79LLLRR61 pKa = 11.84LAGSRR66 pKa = 11.84AEE68 pKa = 3.96ALQEE72 pKa = 3.95EE73 pKa = 5.03VAEE76 pKa = 4.25ALATVTEE83 pKa = 4.86WPCLLEE89 pKa = 4.12YY90 pKa = 10.28TSEE93 pKa = 4.2EE94 pKa = 4.34EE95 pKa = 4.46PPVPLDD101 pKa = 3.59GVSLKK106 pKa = 10.82LSCTYY111 pKa = 10.34PADD114 pKa = 3.67TDD116 pKa = 4.06EE117 pKa = 5.81DD118 pKa = 4.62DD119 pKa = 4.28EE120 pKa = 5.45DD121 pKa = 4.4AVPHH125 pKa = 6.32NIFYY129 pKa = 11.16LEE131 pKa = 3.91GVYY134 pKa = 10.71LRR136 pKa = 11.84IEE138 pKa = 4.02PP139 pKa = 4.07
MM1 pKa = 7.39KK2 pKa = 10.48RR3 pKa = 11.84LIDD6 pKa = 4.37ALADD10 pKa = 3.86DD11 pKa = 4.83ADD13 pKa = 4.09FFVEE17 pKa = 4.74HH18 pKa = 5.67VQITAIVFDD27 pKa = 3.77NTNDD31 pKa = 3.3VTVWATTYY39 pKa = 10.58FDD41 pKa = 3.89EE42 pKa = 4.67EE43 pKa = 4.34LHH45 pKa = 6.19FFHH48 pKa = 7.36LGLQFQYY55 pKa = 11.47LDD57 pKa = 3.79LLLRR61 pKa = 11.84LAGSRR66 pKa = 11.84AEE68 pKa = 3.96ALQEE72 pKa = 3.95EE73 pKa = 5.03VAEE76 pKa = 4.25ALATVTEE83 pKa = 4.86WPCLLEE89 pKa = 4.12YY90 pKa = 10.28TSEE93 pKa = 4.2EE94 pKa = 4.34EE95 pKa = 4.46PPVPLDD101 pKa = 3.59GVSLKK106 pKa = 10.82LSCTYY111 pKa = 10.34PADD114 pKa = 3.67TDD116 pKa = 4.06EE117 pKa = 5.81DD118 pKa = 4.62DD119 pKa = 4.28EE120 pKa = 5.45DD121 pKa = 4.4AVPHH125 pKa = 6.32NIFYY129 pKa = 11.16LEE131 pKa = 3.91GVYY134 pKa = 10.71LRR136 pKa = 11.84IEE138 pKa = 4.02PP139 pKa = 4.07
Molecular weight: 15.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B8A3K3|A0A5B8A3K3_9BACT Omp85 domain-containing protein OS=Hymenobacter jejuensis OX=2502781 GN=FHG12_18210 PE=4 SV=1
MM1 pKa = 7.81DD2 pKa = 3.43TTTIFRR8 pKa = 11.84RR9 pKa = 11.84CVPRR13 pKa = 11.84QRR15 pKa = 11.84VFWVLLVCLFSFFVHH30 pKa = 6.67LGAPEE35 pKa = 3.94VSLMEE40 pKa = 4.09SRR42 pKa = 11.84NFVAARR48 pKa = 11.84EE49 pKa = 4.01MAAGGSWLIPTMNGEE64 pKa = 4.2LRR66 pKa = 11.84LAKK69 pKa = 10.23PPLPTWAVAALLKK82 pKa = 10.89VVGPTQNLAVLRR94 pKa = 11.84LPAALMATLLIFFFWGLARR113 pKa = 11.84EE114 pKa = 4.39LTRR117 pKa = 11.84EE118 pKa = 4.09QPAEE122 pKa = 3.56AAAPGRR128 pKa = 11.84TAWLSALVLGSSLLLVTTGRR148 pKa = 11.84EE149 pKa = 4.3GQWDD153 pKa = 3.47IFSNCFMVGSLWLLVRR169 pKa = 11.84GLNSAGASYY178 pKa = 11.14GRR180 pKa = 11.84FVGAGVLLGCSFLSKK195 pKa = 10.98GPVALFGLLLPFAACYY211 pKa = 9.86CIQRR215 pKa = 11.84LNPAGRR221 pKa = 11.84ARR223 pKa = 11.84LRR225 pKa = 11.84VHH227 pKa = 7.28RR228 pKa = 11.84GGLLVALLIAVLVGGAWPLYY248 pKa = 9.43IFEE251 pKa = 4.85HH252 pKa = 5.38VQPAALAVAQTEE264 pKa = 4.29VTAWRR269 pKa = 11.84EE270 pKa = 3.75RR271 pKa = 11.84HH272 pKa = 4.67VQAWWYY278 pKa = 9.1YY279 pKa = 10.24LNFPVFAGVWAVVALAALAVPYY301 pKa = 10.46ARR303 pKa = 11.84RR304 pKa = 11.84RR305 pKa = 11.84SEE307 pKa = 4.5AYY309 pKa = 8.98IPYY312 pKa = 9.65VFALGWLLVTLVLLSAVPEE331 pKa = 4.06KK332 pKa = 10.63KK333 pKa = 9.59EE334 pKa = 4.53RR335 pKa = 11.84YY336 pKa = 7.77MLPLMPPLALLMGGMLRR353 pKa = 11.84HH354 pKa = 6.28WEE356 pKa = 4.11TSRR359 pKa = 11.84TLPRR363 pKa = 11.84LRR365 pKa = 11.84PADD368 pKa = 4.06GVWLRR373 pKa = 11.84VWAGLLLLVCLLVPVAMVVIKK394 pKa = 10.9LPGFEE399 pKa = 4.13PATLRR404 pKa = 11.84FGAALGVFGSLAVLVWRR421 pKa = 11.84QGWQRR426 pKa = 11.84RR427 pKa = 11.84QPVHH431 pKa = 5.61LTGASAAGMAAVLALLMPAYY451 pKa = 9.16PVWEE455 pKa = 4.01ARR457 pKa = 11.84KK458 pKa = 9.46GDD460 pKa = 3.19GSLRR464 pKa = 11.84HH465 pKa = 6.08ISDD468 pKa = 3.0VRR470 pKa = 11.84EE471 pKa = 3.79RR472 pKa = 11.84AEE474 pKa = 4.04LQRR477 pKa = 11.84LPWVGQQEE485 pKa = 4.26VHH487 pKa = 6.95IKK489 pKa = 9.41QVWNAGRR496 pKa = 11.84SIPTWHH502 pKa = 7.81PDD504 pKa = 2.79TTAAEE509 pKa = 4.38LPSLPVVVFSSDD521 pKa = 3.12PHH523 pKa = 7.48VLEE526 pKa = 4.09KK527 pKa = 10.89LPRR530 pKa = 11.84KK531 pKa = 8.34WRR533 pKa = 11.84ARR535 pKa = 11.84LRR537 pKa = 11.84SEE539 pKa = 5.02RR540 pKa = 11.84IDD542 pKa = 3.48SFFLGRR548 pKa = 11.84TAKK551 pKa = 10.65DD552 pKa = 3.5GWWMVSLLQRR562 pKa = 11.84KK563 pKa = 7.97VNN565 pKa = 3.53
MM1 pKa = 7.81DD2 pKa = 3.43TTTIFRR8 pKa = 11.84RR9 pKa = 11.84CVPRR13 pKa = 11.84QRR15 pKa = 11.84VFWVLLVCLFSFFVHH30 pKa = 6.67LGAPEE35 pKa = 3.94VSLMEE40 pKa = 4.09SRR42 pKa = 11.84NFVAARR48 pKa = 11.84EE49 pKa = 4.01MAAGGSWLIPTMNGEE64 pKa = 4.2LRR66 pKa = 11.84LAKK69 pKa = 10.23PPLPTWAVAALLKK82 pKa = 10.89VVGPTQNLAVLRR94 pKa = 11.84LPAALMATLLIFFFWGLARR113 pKa = 11.84EE114 pKa = 4.39LTRR117 pKa = 11.84EE118 pKa = 4.09QPAEE122 pKa = 3.56AAAPGRR128 pKa = 11.84TAWLSALVLGSSLLLVTTGRR148 pKa = 11.84EE149 pKa = 4.3GQWDD153 pKa = 3.47IFSNCFMVGSLWLLVRR169 pKa = 11.84GLNSAGASYY178 pKa = 11.14GRR180 pKa = 11.84FVGAGVLLGCSFLSKK195 pKa = 10.98GPVALFGLLLPFAACYY211 pKa = 9.86CIQRR215 pKa = 11.84LNPAGRR221 pKa = 11.84ARR223 pKa = 11.84LRR225 pKa = 11.84VHH227 pKa = 7.28RR228 pKa = 11.84GGLLVALLIAVLVGGAWPLYY248 pKa = 9.43IFEE251 pKa = 4.85HH252 pKa = 5.38VQPAALAVAQTEE264 pKa = 4.29VTAWRR269 pKa = 11.84EE270 pKa = 3.75RR271 pKa = 11.84HH272 pKa = 4.67VQAWWYY278 pKa = 9.1YY279 pKa = 10.24LNFPVFAGVWAVVALAALAVPYY301 pKa = 10.46ARR303 pKa = 11.84RR304 pKa = 11.84RR305 pKa = 11.84SEE307 pKa = 4.5AYY309 pKa = 8.98IPYY312 pKa = 9.65VFALGWLLVTLVLLSAVPEE331 pKa = 4.06KK332 pKa = 10.63KK333 pKa = 9.59EE334 pKa = 4.53RR335 pKa = 11.84YY336 pKa = 7.77MLPLMPPLALLMGGMLRR353 pKa = 11.84HH354 pKa = 6.28WEE356 pKa = 4.11TSRR359 pKa = 11.84TLPRR363 pKa = 11.84LRR365 pKa = 11.84PADD368 pKa = 4.06GVWLRR373 pKa = 11.84VWAGLLLLVCLLVPVAMVVIKK394 pKa = 10.9LPGFEE399 pKa = 4.13PATLRR404 pKa = 11.84FGAALGVFGSLAVLVWRR421 pKa = 11.84QGWQRR426 pKa = 11.84RR427 pKa = 11.84QPVHH431 pKa = 5.61LTGASAAGMAAVLALLMPAYY451 pKa = 9.16PVWEE455 pKa = 4.01ARR457 pKa = 11.84KK458 pKa = 9.46GDD460 pKa = 3.19GSLRR464 pKa = 11.84HH465 pKa = 6.08ISDD468 pKa = 3.0VRR470 pKa = 11.84EE471 pKa = 3.79RR472 pKa = 11.84AEE474 pKa = 4.04LQRR477 pKa = 11.84LPWVGQQEE485 pKa = 4.26VHH487 pKa = 6.95IKK489 pKa = 9.41QVWNAGRR496 pKa = 11.84SIPTWHH502 pKa = 7.81PDD504 pKa = 2.79TTAAEE509 pKa = 4.38LPSLPVVVFSSDD521 pKa = 3.12PHH523 pKa = 7.48VLEE526 pKa = 4.09KK527 pKa = 10.89LPRR530 pKa = 11.84KK531 pKa = 8.34WRR533 pKa = 11.84ARR535 pKa = 11.84LRR537 pKa = 11.84SEE539 pKa = 5.02RR540 pKa = 11.84IDD542 pKa = 3.48SFFLGRR548 pKa = 11.84TAKK551 pKa = 10.65DD552 pKa = 3.5GWWMVSLLQRR562 pKa = 11.84KK563 pKa = 7.97VNN565 pKa = 3.53
Molecular weight: 62.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1465612 |
25 |
2462 |
357.5 |
39.56 |
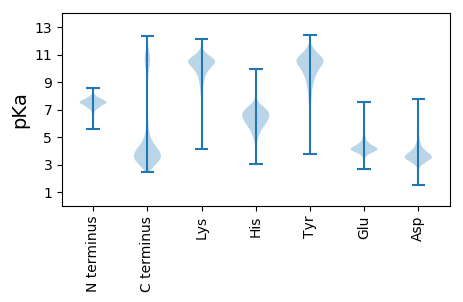
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.882 ± 0.045 | 0.721 ± 0.01 |
5.034 ± 0.026 | 5.336 ± 0.04 |
4.256 ± 0.021 | 7.386 ± 0.036 |
2.108 ± 0.022 | 4.843 ± 0.031 |
4.461 ± 0.034 | 10.593 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.968 ± 0.018 | 4.055 ± 0.035 |
4.999 ± 0.026 | 4.624 ± 0.028 |
5.767 ± 0.032 | 5.737 ± 0.03 |
6.164 ± 0.047 | 7.181 ± 0.028 |
1.266 ± 0.016 | 3.617 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
