
Luffa aphid-borne yellows virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus; unclassified Polerovirus
Average proteome isoelectric point is 7.68
Get precalculated fractions of proteins
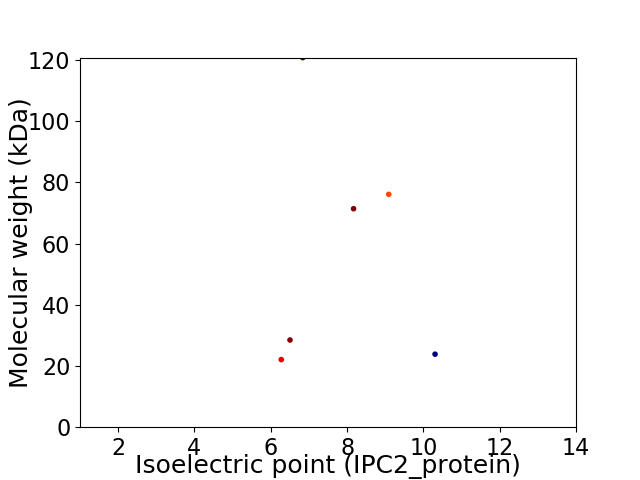
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
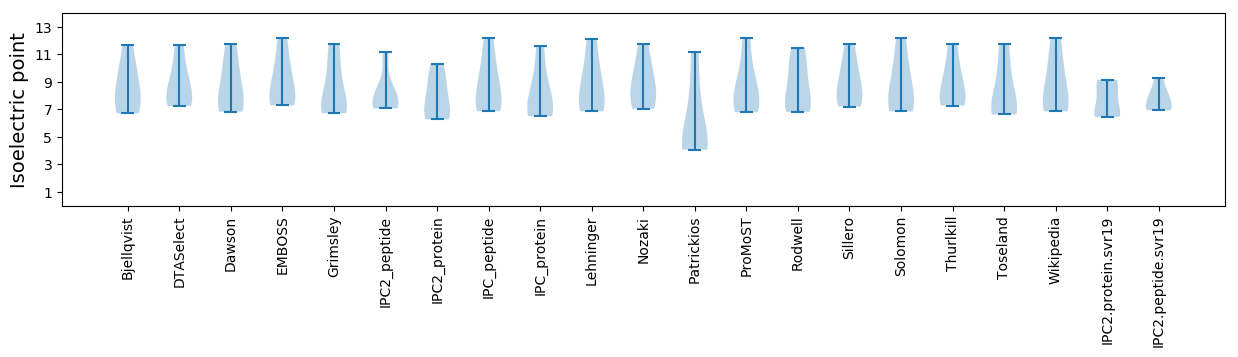
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W8JWL6|W8JWL6_9LUTE Movement protein OS=Luffa aphid-borne yellows virus OX=1462682 PE=3 SV=2
MM1 pKa = 7.17GHH3 pKa = 6.19MLRR6 pKa = 11.84VGHH9 pKa = 6.51SGTGDD14 pKa = 3.2VDD16 pKa = 3.39NDD18 pKa = 3.34EE19 pKa = 4.14PRR21 pKa = 11.84EE22 pKa = 3.98YY23 pKa = 11.35GMSSHH28 pKa = 5.88QAAVQSWSSLTYY40 pKa = 10.77NPDD43 pKa = 2.76VDD45 pKa = 4.57EE46 pKa = 5.66IDD48 pKa = 3.87VIGEE52 pKa = 3.98EE53 pKa = 4.47EE54 pKa = 4.12EE55 pKa = 4.66VEE57 pKa = 4.34FPEE60 pKa = 4.57VQGLLKK66 pKa = 10.71SSSLYY71 pKa = 9.53YY72 pKa = 8.68QTSRR76 pKa = 11.84EE77 pKa = 4.06TXPGQSSSDD86 pKa = 3.78HH87 pKa = 6.09LCHH90 pKa = 7.29RR91 pKa = 11.84SQNLRR96 pKa = 11.84QEE98 pKa = 4.36YY99 pKa = 10.42SSPTLAIRR107 pKa = 11.84YY108 pKa = 7.51HH109 pKa = 6.71RR110 pKa = 11.84SHH112 pKa = 7.1SVFAPRR118 pKa = 11.84RR119 pKa = 11.84LLPMVAPSSISSIPLALQRR138 pKa = 11.84NSTRR142 pKa = 11.84SSIGSRR148 pKa = 11.84YY149 pKa = 7.1PTEE152 pKa = 4.01PLKK155 pKa = 10.11MLRR158 pKa = 11.84GEE160 pKa = 4.08GHH162 pKa = 6.49RR163 pKa = 11.84SEE165 pKa = 4.75EE166 pKa = 4.03RR167 pKa = 11.84SGTPRR172 pKa = 11.84AQSNSGSYY180 pKa = 8.24TKK182 pKa = 10.81EE183 pKa = 3.45MAKK186 pKa = 9.98QKK188 pKa = 10.77SLARR192 pKa = 11.84SPSQWWW198 pKa = 3.06
MM1 pKa = 7.17GHH3 pKa = 6.19MLRR6 pKa = 11.84VGHH9 pKa = 6.51SGTGDD14 pKa = 3.2VDD16 pKa = 3.39NDD18 pKa = 3.34EE19 pKa = 4.14PRR21 pKa = 11.84EE22 pKa = 3.98YY23 pKa = 11.35GMSSHH28 pKa = 5.88QAAVQSWSSLTYY40 pKa = 10.77NPDD43 pKa = 2.76VDD45 pKa = 4.57EE46 pKa = 5.66IDD48 pKa = 3.87VIGEE52 pKa = 3.98EE53 pKa = 4.47EE54 pKa = 4.12EE55 pKa = 4.66VEE57 pKa = 4.34FPEE60 pKa = 4.57VQGLLKK66 pKa = 10.71SSSLYY71 pKa = 9.53YY72 pKa = 8.68QTSRR76 pKa = 11.84EE77 pKa = 4.06TXPGQSSSDD86 pKa = 3.78HH87 pKa = 6.09LCHH90 pKa = 7.29RR91 pKa = 11.84SQNLRR96 pKa = 11.84QEE98 pKa = 4.36YY99 pKa = 10.42SSPTLAIRR107 pKa = 11.84YY108 pKa = 7.51HH109 pKa = 6.71RR110 pKa = 11.84SHH112 pKa = 7.1SVFAPRR118 pKa = 11.84RR119 pKa = 11.84LLPMVAPSSISSIPLALQRR138 pKa = 11.84NSTRR142 pKa = 11.84SSIGSRR148 pKa = 11.84YY149 pKa = 7.1PTEE152 pKa = 4.01PLKK155 pKa = 10.11MLRR158 pKa = 11.84GEE160 pKa = 4.08GHH162 pKa = 6.49RR163 pKa = 11.84SEE165 pKa = 4.75EE166 pKa = 4.03RR167 pKa = 11.84SGTPRR172 pKa = 11.84AQSNSGSYY180 pKa = 8.24TKK182 pKa = 10.81EE183 pKa = 3.45MAKK186 pKa = 9.98QKK188 pKa = 10.77SLARR192 pKa = 11.84SPSQWWW198 pKa = 3.06
Molecular weight: 22.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8JD48|W8JD48_9LUTE Serine protease OS=Luffa aphid-borne yellows virus OX=1462682 PE=4 SV=2
MM1 pKa = 6.6NTAGGRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84NGSYY13 pKa = 8.59VTSRR17 pKa = 11.84TLRR20 pKa = 11.84NRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84QRR27 pKa = 11.84RR28 pKa = 11.84AARR31 pKa = 11.84VRR33 pKa = 11.84NVIPPGGGPIMVLPNLQSRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84NRR57 pKa = 11.84RR58 pKa = 11.84NRR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84GGGVSRR69 pKa = 11.84GSGSTEE75 pKa = 3.55VFKK78 pKa = 11.12FILPDD83 pKa = 3.65LKK85 pKa = 11.09GNSAGTIKK93 pKa = 10.66FGPSLSQKK101 pKa = 10.45PEE103 pKa = 3.57FATGILKK110 pKa = 10.48SYY112 pKa = 10.74ARR114 pKa = 11.84YY115 pKa = 9.76KK116 pKa = 10.27ISQVTLSFRR125 pKa = 11.84SEE127 pKa = 3.75ASATDD132 pKa = 3.64GGALVYY138 pKa = 10.74QLDD141 pKa = 4.17TTCAATQLDD150 pKa = 3.84SKK152 pKa = 10.62LYY154 pKa = 10.27RR155 pKa = 11.84FSISNRR161 pKa = 11.84APQNVTWRR169 pKa = 11.84GAQIRR174 pKa = 11.84GEE176 pKa = 4.27EE177 pKa = 4.01WHH179 pKa = 6.5STSSEE184 pKa = 4.27QFWVLYY190 pKa = 9.6QGNGEE195 pKa = 4.18AKK197 pKa = 8.83VAGSITVTMVVNFLDD212 pKa = 4.25PKK214 pKa = 10.73
MM1 pKa = 6.6NTAGGRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84NGSYY13 pKa = 8.59VTSRR17 pKa = 11.84TLRR20 pKa = 11.84NRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84QRR27 pKa = 11.84RR28 pKa = 11.84AARR31 pKa = 11.84VRR33 pKa = 11.84NVIPPGGGPIMVLPNLQSRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84NRR57 pKa = 11.84RR58 pKa = 11.84NRR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84GGGVSRR69 pKa = 11.84GSGSTEE75 pKa = 3.55VFKK78 pKa = 11.12FILPDD83 pKa = 3.65LKK85 pKa = 11.09GNSAGTIKK93 pKa = 10.66FGPSLSQKK101 pKa = 10.45PEE103 pKa = 3.57FATGILKK110 pKa = 10.48SYY112 pKa = 10.74ARR114 pKa = 11.84YY115 pKa = 9.76KK116 pKa = 10.27ISQVTLSFRR125 pKa = 11.84SEE127 pKa = 3.75ASATDD132 pKa = 3.64GGALVYY138 pKa = 10.74QLDD141 pKa = 4.17TTCAATQLDD150 pKa = 3.84SKK152 pKa = 10.62LYY154 pKa = 10.27RR155 pKa = 11.84FSISNRR161 pKa = 11.84APQNVTWRR169 pKa = 11.84GAQIRR174 pKa = 11.84GEE176 pKa = 4.27EE177 pKa = 4.01WHH179 pKa = 6.5STSSEE184 pKa = 4.27QFWVLYY190 pKa = 9.6QGNGEE195 pKa = 4.18AKK197 pKa = 8.83VAGSITVTMVVNFLDD212 pKa = 4.25PKK214 pKa = 10.73
Molecular weight: 23.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3092 |
198 |
1082 |
515.3 |
57.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.05 ± 0.367 | 2.005 ± 0.565 |
4.107 ± 0.383 | 5.595 ± 0.426 |
3.784 ± 0.353 | 7.503 ± 0.517 |
2.426 ± 0.412 | 3.913 ± 0.474 |
5.045 ± 0.682 | 9.799 ± 1.098 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.714 ± 0.219 | 3.655 ± 0.277 |
6.339 ± 0.514 | 3.816 ± 0.192 |
6.824 ± 1.374 | 9.541 ± 0.782 |
5.563 ± 0.384 | 6.21 ± 0.261 |
1.94 ± 0.173 | 2.943 ± 0.121 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
