
Shahe hepe-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
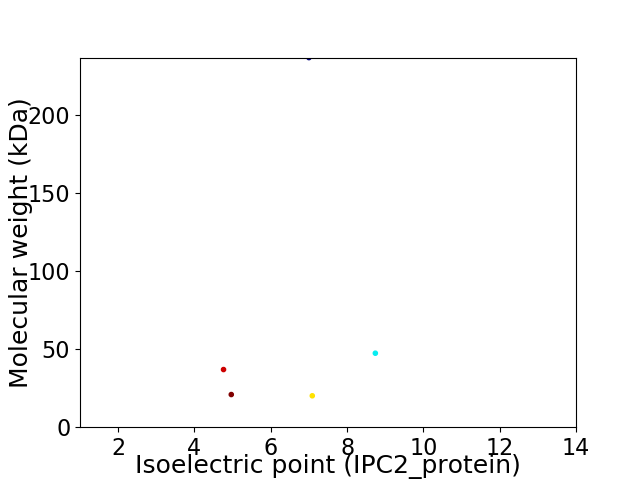
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KK16|A0A1L3KK16_9VIRU Uncharacterized protein OS=Shahe hepe-like virus 1 OX=1923415 PE=4 SV=1
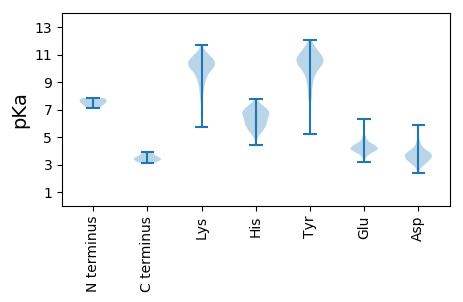
MM1 pKa = 7.42NSDD4 pKa = 3.66PAQANSVSEE13 pKa = 4.1GAPVEE18 pKa = 4.17LSTGSTGLPSSDD30 pKa = 3.37FTDD33 pKa = 3.5SKK35 pKa = 11.33PLFLEE40 pKa = 5.07AGTQHH45 pKa = 6.17TAVHH49 pKa = 6.76CFRR52 pKa = 11.84VAKK55 pKa = 10.58DD56 pKa = 3.64LVTLAEE62 pKa = 4.38GSKK65 pKa = 10.31PGEE68 pKa = 3.83ICFVQPLTPFGIDD81 pKa = 3.03NTATNLASDD90 pKa = 3.75YY91 pKa = 10.75EE92 pKa = 4.56YY93 pKa = 11.6YY94 pKa = 10.66SFKK97 pKa = 10.77NVEE100 pKa = 3.82IAIEE104 pKa = 4.08AVSPFGTSSGGMQVAWITDD123 pKa = 3.85PEE125 pKa = 4.58NVTLPQGDD133 pKa = 4.54DD134 pKa = 3.75GQKK137 pKa = 8.95LTSLAKK143 pKa = 10.33VIRR146 pKa = 11.84QDD148 pKa = 2.89GSVLVRR154 pKa = 11.84PRR156 pKa = 11.84NSALFNISTEE166 pKa = 4.05GARR169 pKa = 11.84YY170 pKa = 8.94CLPGTEE176 pKa = 4.53SRR178 pKa = 11.84LSSFGSLVALVRR190 pKa = 11.84SPPGTGDD197 pKa = 3.31VADD200 pKa = 5.37FAATITGEE208 pKa = 3.85IHH210 pKa = 7.04FMRR213 pKa = 11.84STRR216 pKa = 11.84NAPVYY221 pKa = 10.25IGEE224 pKa = 4.22FPIGTFTIDD233 pKa = 3.08EE234 pKa = 4.36TQPLIEE240 pKa = 4.84EE241 pKa = 4.46NDD243 pKa = 4.08LILPVRR249 pKa = 11.84LSSNDD254 pKa = 3.09VQTDD258 pKa = 2.89ISRR261 pKa = 11.84WDD263 pKa = 3.2FRR265 pKa = 11.84TPLKK269 pKa = 9.99LTLRR273 pKa = 11.84GKK275 pKa = 10.21DD276 pKa = 3.37GRR278 pKa = 11.84FTKK281 pKa = 10.04IHH283 pKa = 6.58EE284 pKa = 4.36IKK286 pKa = 10.8ADD288 pKa = 3.55FATFKK293 pKa = 9.93LTNIDD298 pKa = 3.29DD299 pKa = 3.96QYY301 pKa = 12.05AEE303 pKa = 5.13GILMIDD309 pKa = 3.78LPFYY313 pKa = 11.1GLTNPVVRR321 pKa = 11.84INSFPRR327 pKa = 11.84QIINSYY333 pKa = 10.07EE334 pKa = 3.92VLRR337 pKa = 11.84NN338 pKa = 3.42
MM1 pKa = 7.42NSDD4 pKa = 3.66PAQANSVSEE13 pKa = 4.1GAPVEE18 pKa = 4.17LSTGSTGLPSSDD30 pKa = 3.37FTDD33 pKa = 3.5SKK35 pKa = 11.33PLFLEE40 pKa = 5.07AGTQHH45 pKa = 6.17TAVHH49 pKa = 6.76CFRR52 pKa = 11.84VAKK55 pKa = 10.58DD56 pKa = 3.64LVTLAEE62 pKa = 4.38GSKK65 pKa = 10.31PGEE68 pKa = 3.83ICFVQPLTPFGIDD81 pKa = 3.03NTATNLASDD90 pKa = 3.75YY91 pKa = 10.75EE92 pKa = 4.56YY93 pKa = 11.6YY94 pKa = 10.66SFKK97 pKa = 10.77NVEE100 pKa = 3.82IAIEE104 pKa = 4.08AVSPFGTSSGGMQVAWITDD123 pKa = 3.85PEE125 pKa = 4.58NVTLPQGDD133 pKa = 4.54DD134 pKa = 3.75GQKK137 pKa = 8.95LTSLAKK143 pKa = 10.33VIRR146 pKa = 11.84QDD148 pKa = 2.89GSVLVRR154 pKa = 11.84PRR156 pKa = 11.84NSALFNISTEE166 pKa = 4.05GARR169 pKa = 11.84YY170 pKa = 8.94CLPGTEE176 pKa = 4.53SRR178 pKa = 11.84LSSFGSLVALVRR190 pKa = 11.84SPPGTGDD197 pKa = 3.31VADD200 pKa = 5.37FAATITGEE208 pKa = 3.85IHH210 pKa = 7.04FMRR213 pKa = 11.84STRR216 pKa = 11.84NAPVYY221 pKa = 10.25IGEE224 pKa = 4.22FPIGTFTIDD233 pKa = 3.08EE234 pKa = 4.36TQPLIEE240 pKa = 4.84EE241 pKa = 4.46NDD243 pKa = 4.08LILPVRR249 pKa = 11.84LSSNDD254 pKa = 3.09VQTDD258 pKa = 2.89ISRR261 pKa = 11.84WDD263 pKa = 3.2FRR265 pKa = 11.84TPLKK269 pKa = 9.99LTLRR273 pKa = 11.84GKK275 pKa = 10.21DD276 pKa = 3.37GRR278 pKa = 11.84FTKK281 pKa = 10.04IHH283 pKa = 6.58EE284 pKa = 4.36IKK286 pKa = 10.8ADD288 pKa = 3.55FATFKK293 pKa = 9.93LTNIDD298 pKa = 3.29DD299 pKa = 3.96QYY301 pKa = 12.05AEE303 pKa = 5.13GILMIDD309 pKa = 3.78LPFYY313 pKa = 11.1GLTNPVVRR321 pKa = 11.84INSFPRR327 pKa = 11.84QIINSYY333 pKa = 10.07EE334 pKa = 3.92VLRR337 pKa = 11.84NN338 pKa = 3.42
Molecular weight: 36.88 kDa
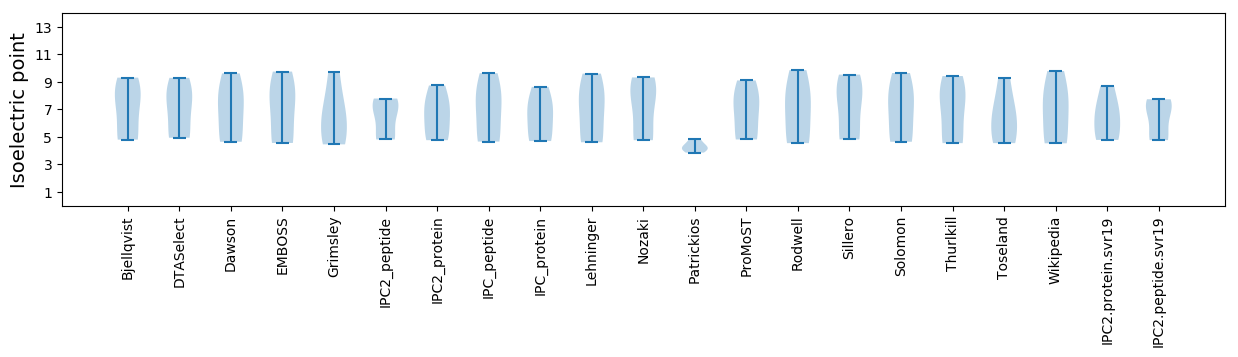
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KK10|A0A1L3KK10_9VIRU Putative structural protein OS=Shahe hepe-like virus 1 OX=1923415 PE=4 SV=1
MM1 pKa = 7.11QVRR4 pKa = 11.84ILTQYY9 pKa = 10.26QRR11 pKa = 11.84EE12 pKa = 4.02QRR14 pKa = 11.84KK15 pKa = 9.84DD16 pKa = 3.08RR17 pKa = 11.84VKK19 pKa = 11.24YY20 pKa = 10.45LLQTFKK26 pKa = 11.17LSLSKK31 pKa = 10.92VPKK34 pKa = 9.43TDD36 pKa = 3.7LFRR39 pKa = 11.84QINTIISNQFKK50 pKa = 9.59MRR52 pKa = 11.84PLMPVPFSVTPPKK65 pKa = 10.05QASHH69 pKa = 6.94SIHH72 pKa = 6.65HH73 pKa = 5.81SRR75 pKa = 11.84VRR77 pKa = 11.84NTGLAKK83 pKa = 10.96LITNGKK89 pKa = 7.97NDD91 pKa = 3.47EE92 pKa = 4.5FEE94 pKa = 4.16FHH96 pKa = 7.12LLDD99 pKa = 4.93AEE101 pKa = 4.17NKK103 pKa = 10.19YY104 pKa = 10.85FFDD107 pKa = 3.91EE108 pKa = 4.24NALEE112 pKa = 3.97MFKK115 pKa = 10.49NHH117 pKa = 5.42EE118 pKa = 4.17YY119 pKa = 11.07YY120 pKa = 10.31RR121 pKa = 11.84IKK123 pKa = 10.3EE124 pKa = 3.57ITMTINNTEE133 pKa = 3.33NWLKK137 pKa = 10.75VRR139 pKa = 11.84GAFQASYY146 pKa = 10.57VHH148 pKa = 6.98DD149 pKa = 4.23PEE151 pKa = 6.31GRR153 pKa = 11.84ADD155 pKa = 3.71PLSVRR160 pKa = 11.84FQDD163 pKa = 3.62TQLVRR168 pKa = 11.84IGKK171 pKa = 9.37AVVWQIPVLKK181 pKa = 9.68KK182 pKa = 9.67WLYY185 pKa = 5.22TTSGGSSGSKK195 pKa = 9.76RR196 pKa = 11.84LSAAGKK202 pKa = 9.43VIIKK206 pKa = 9.92IRR208 pKa = 11.84SKK210 pKa = 10.86QNNVKK215 pKa = 10.29DD216 pKa = 3.92APPNGAEE223 pKa = 4.14SYY225 pKa = 10.2KK226 pKa = 10.72VAVTIDD232 pKa = 3.99AIIEE236 pKa = 4.36FCSKK240 pKa = 9.23TRR242 pKa = 11.84ILRR245 pKa = 11.84LGGIIYY251 pKa = 8.79RR252 pKa = 11.84IPFTGDD258 pKa = 2.37AGGQVVNDD266 pKa = 3.51YY267 pKa = 10.94VRR269 pKa = 11.84LFTKK273 pKa = 10.29TDD275 pKa = 3.17DD276 pKa = 3.39RR277 pKa = 11.84DD278 pKa = 3.61YY279 pKa = 11.65YY280 pKa = 11.24LDD282 pKa = 3.35IRR284 pKa = 11.84IAEE287 pKa = 4.59LFPFKK292 pKa = 10.07PAQGFFFLQSPVMVTFKK309 pKa = 10.43FHH311 pKa = 7.76SSEE314 pKa = 4.06EE315 pKa = 4.18TNKK318 pKa = 10.38IIIYY322 pKa = 10.36DD323 pKa = 3.77FMMTNGNFYY332 pKa = 10.42MNEE335 pKa = 3.92DD336 pKa = 3.98SLSIWLRR343 pKa = 11.84TKK345 pKa = 10.69VDD347 pKa = 2.72IGSAYY352 pKa = 10.54SGTDD356 pKa = 3.48FQPDD360 pKa = 3.24EE361 pKa = 4.44LSIEE365 pKa = 4.09FQYY368 pKa = 11.57DD369 pKa = 3.09SAYY372 pKa = 10.57LEE374 pKa = 4.0QGIIPNFITFMPDD387 pKa = 2.82DD388 pKa = 3.92FEE390 pKa = 4.99KK391 pKa = 10.96EE392 pKa = 3.87KK393 pKa = 10.42TFSYY397 pKa = 10.42RR398 pKa = 11.84DD399 pKa = 3.2QRR401 pKa = 11.84PRR403 pKa = 11.84KK404 pKa = 9.01FLQQ407 pKa = 3.4
MM1 pKa = 7.11QVRR4 pKa = 11.84ILTQYY9 pKa = 10.26QRR11 pKa = 11.84EE12 pKa = 4.02QRR14 pKa = 11.84KK15 pKa = 9.84DD16 pKa = 3.08RR17 pKa = 11.84VKK19 pKa = 11.24YY20 pKa = 10.45LLQTFKK26 pKa = 11.17LSLSKK31 pKa = 10.92VPKK34 pKa = 9.43TDD36 pKa = 3.7LFRR39 pKa = 11.84QINTIISNQFKK50 pKa = 9.59MRR52 pKa = 11.84PLMPVPFSVTPPKK65 pKa = 10.05QASHH69 pKa = 6.94SIHH72 pKa = 6.65HH73 pKa = 5.81SRR75 pKa = 11.84VRR77 pKa = 11.84NTGLAKK83 pKa = 10.96LITNGKK89 pKa = 7.97NDD91 pKa = 3.47EE92 pKa = 4.5FEE94 pKa = 4.16FHH96 pKa = 7.12LLDD99 pKa = 4.93AEE101 pKa = 4.17NKK103 pKa = 10.19YY104 pKa = 10.85FFDD107 pKa = 3.91EE108 pKa = 4.24NALEE112 pKa = 3.97MFKK115 pKa = 10.49NHH117 pKa = 5.42EE118 pKa = 4.17YY119 pKa = 11.07YY120 pKa = 10.31RR121 pKa = 11.84IKK123 pKa = 10.3EE124 pKa = 3.57ITMTINNTEE133 pKa = 3.33NWLKK137 pKa = 10.75VRR139 pKa = 11.84GAFQASYY146 pKa = 10.57VHH148 pKa = 6.98DD149 pKa = 4.23PEE151 pKa = 6.31GRR153 pKa = 11.84ADD155 pKa = 3.71PLSVRR160 pKa = 11.84FQDD163 pKa = 3.62TQLVRR168 pKa = 11.84IGKK171 pKa = 9.37AVVWQIPVLKK181 pKa = 9.68KK182 pKa = 9.67WLYY185 pKa = 5.22TTSGGSSGSKK195 pKa = 9.76RR196 pKa = 11.84LSAAGKK202 pKa = 9.43VIIKK206 pKa = 9.92IRR208 pKa = 11.84SKK210 pKa = 10.86QNNVKK215 pKa = 10.29DD216 pKa = 3.92APPNGAEE223 pKa = 4.14SYY225 pKa = 10.2KK226 pKa = 10.72VAVTIDD232 pKa = 3.99AIIEE236 pKa = 4.36FCSKK240 pKa = 9.23TRR242 pKa = 11.84ILRR245 pKa = 11.84LGGIIYY251 pKa = 8.79RR252 pKa = 11.84IPFTGDD258 pKa = 2.37AGGQVVNDD266 pKa = 3.51YY267 pKa = 10.94VRR269 pKa = 11.84LFTKK273 pKa = 10.29TDD275 pKa = 3.17DD276 pKa = 3.39RR277 pKa = 11.84DD278 pKa = 3.61YY279 pKa = 11.65YY280 pKa = 11.24LDD282 pKa = 3.35IRR284 pKa = 11.84IAEE287 pKa = 4.59LFPFKK292 pKa = 10.07PAQGFFFLQSPVMVTFKK309 pKa = 10.43FHH311 pKa = 7.76SSEE314 pKa = 4.06EE315 pKa = 4.18TNKK318 pKa = 10.38IIIYY322 pKa = 10.36DD323 pKa = 3.77FMMTNGNFYY332 pKa = 10.42MNEE335 pKa = 3.92DD336 pKa = 3.98SLSIWLRR343 pKa = 11.84TKK345 pKa = 10.69VDD347 pKa = 2.72IGSAYY352 pKa = 10.54SGTDD356 pKa = 3.48FQPDD360 pKa = 3.24EE361 pKa = 4.44LSIEE365 pKa = 4.09FQYY368 pKa = 11.57DD369 pKa = 3.09SAYY372 pKa = 10.57LEE374 pKa = 4.0QGIIPNFITFMPDD387 pKa = 2.82DD388 pKa = 3.92FEE390 pKa = 4.99KK391 pKa = 10.96EE392 pKa = 3.87KK393 pKa = 10.42TFSYY397 pKa = 10.42RR398 pKa = 11.84DD399 pKa = 3.2QRR401 pKa = 11.84PRR403 pKa = 11.84KK404 pKa = 9.01FLQQ407 pKa = 3.4
Molecular weight: 47.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3206 |
173 |
2103 |
641.2 |
72.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.174 ± 0.938 | 1.372 ± 0.514 |
5.646 ± 0.309 | 6.02 ± 0.425 |
5.552 ± 0.7 | 5.209 ± 0.518 |
2.277 ± 0.589 | 7.143 ± 0.468 |
7.049 ± 0.957 | 8.141 ± 0.599 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.183 ± 0.314 | 5.739 ± 0.411 |
4.46 ± 0.502 | 3.587 ± 0.619 |
4.523 ± 0.446 | 6.644 ± 0.632 |
7.299 ± 0.74 | 5.77 ± 0.437 |
0.967 ± 0.227 | 3.244 ± 0.31 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
