
Ruegeria phage vB_RpoMi-V15
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.67
Get precalculated fractions of proteins
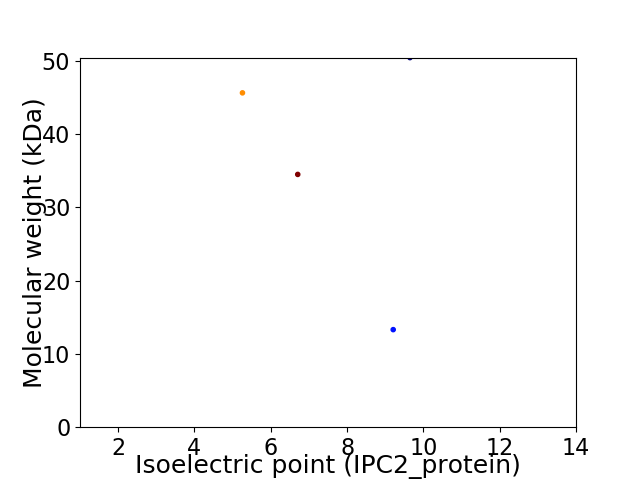
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z4QGJ3|A0A2Z4QGJ3_9VIRU Capsid protein OS=Ruegeria phage vB_RpoMi-V15 OX=2218610 GN=vBRpoMiV15_04 PE=4 SV=1
MM1 pKa = 7.53LANLIMQQGGAKK13 pKa = 9.73SKK15 pKa = 10.48RR16 pKa = 11.84KK17 pKa = 9.33NRR19 pKa = 11.84NPVFTLGGTIKK30 pKa = 10.74PFGLYY35 pKa = 10.08PMFIHH40 pKa = 6.72PVLPGEE46 pKa = 4.31TLNSFTMKK54 pKa = 9.98VHH56 pKa = 6.91LASEE60 pKa = 4.17PLKK63 pKa = 11.19NPLNGAWWEE72 pKa = 3.95TWLFYY77 pKa = 11.39VKK79 pKa = 10.73LNDD82 pKa = 4.26LGPDD86 pKa = 3.45LVEE89 pKa = 4.22MFIKK93 pKa = 10.73DD94 pKa = 3.7GVSTTGHH101 pKa = 5.94TAGSSDD107 pKa = 3.21QLFFTKK113 pKa = 10.18SGQIDD118 pKa = 3.88WVRR121 pKa = 11.84KK122 pKa = 8.48CTDD125 pKa = 3.88LVHH128 pKa = 5.74SHH130 pKa = 5.77YY131 pKa = 10.46FIHH134 pKa = 7.04EE135 pKa = 4.56DD136 pKa = 3.04EE137 pKa = 4.4TARR140 pKa = 11.84TINGVPQVKK149 pKa = 10.07LNSKK153 pKa = 9.29GWWQNLMQNDD163 pKa = 3.35ADD165 pKa = 4.04AAVPTGDD172 pKa = 3.57AADD175 pKa = 4.21MYY177 pKa = 11.75AHH179 pKa = 6.43LQEE182 pKa = 4.6FAILQQMGMQEE193 pKa = 3.97MTYY196 pKa = 10.42EE197 pKa = 4.31KK198 pKa = 10.65YY199 pKa = 10.92LEE201 pKa = 4.34LFGGRR206 pKa = 11.84AAAAGQGKK214 pKa = 9.13PEE216 pKa = 4.38LLLYY220 pKa = 9.42DD221 pKa = 3.99QRR223 pKa = 11.84WKK225 pKa = 11.06KK226 pKa = 9.02PANTINPADD235 pKa = 4.1GTPSSAWVWNEE246 pKa = 3.45EE247 pKa = 4.17TKK249 pKa = 10.9LGSKK253 pKa = 9.82NKK255 pKa = 9.93RR256 pKa = 11.84FNEE259 pKa = 4.08PGFVIAFQTIRR270 pKa = 11.84PKK272 pKa = 10.15MFNGAVRR279 pKa = 11.84TSLVGNMWGFKK290 pKa = 9.68DD291 pKa = 3.78WYY293 pKa = 9.35PAYY296 pKa = 9.68TLDD299 pKa = 5.74DD300 pKa = 3.51PTATVKK306 pKa = 10.07TIANNDD312 pKa = 3.65PVFEE316 pKa = 4.3ASAYY320 pKa = 5.27TTGGTEE326 pKa = 5.02LIYY329 pKa = 10.97DD330 pKa = 4.11HH331 pKa = 7.49SDD333 pKa = 2.99LLAQGEE339 pKa = 4.35QFINDD344 pKa = 3.67YY345 pKa = 10.4APPYY349 pKa = 10.15ALPLSSGMVVEE360 pKa = 5.19GGGTPEE366 pKa = 4.06SFRR369 pKa = 11.84GEE371 pKa = 4.44YY372 pKa = 8.24CTSADD377 pKa = 3.69VDD379 pKa = 4.08ALFQDD384 pKa = 4.5GAKK387 pKa = 9.13TRR389 pKa = 11.84AFYY392 pKa = 10.96DD393 pKa = 4.79GIAFCNILGHH403 pKa = 6.09VADD406 pKa = 4.43NTPLKK411 pKa = 10.84
MM1 pKa = 7.53LANLIMQQGGAKK13 pKa = 9.73SKK15 pKa = 10.48RR16 pKa = 11.84KK17 pKa = 9.33NRR19 pKa = 11.84NPVFTLGGTIKK30 pKa = 10.74PFGLYY35 pKa = 10.08PMFIHH40 pKa = 6.72PVLPGEE46 pKa = 4.31TLNSFTMKK54 pKa = 9.98VHH56 pKa = 6.91LASEE60 pKa = 4.17PLKK63 pKa = 11.19NPLNGAWWEE72 pKa = 3.95TWLFYY77 pKa = 11.39VKK79 pKa = 10.73LNDD82 pKa = 4.26LGPDD86 pKa = 3.45LVEE89 pKa = 4.22MFIKK93 pKa = 10.73DD94 pKa = 3.7GVSTTGHH101 pKa = 5.94TAGSSDD107 pKa = 3.21QLFFTKK113 pKa = 10.18SGQIDD118 pKa = 3.88WVRR121 pKa = 11.84KK122 pKa = 8.48CTDD125 pKa = 3.88LVHH128 pKa = 5.74SHH130 pKa = 5.77YY131 pKa = 10.46FIHH134 pKa = 7.04EE135 pKa = 4.56DD136 pKa = 3.04EE137 pKa = 4.4TARR140 pKa = 11.84TINGVPQVKK149 pKa = 10.07LNSKK153 pKa = 9.29GWWQNLMQNDD163 pKa = 3.35ADD165 pKa = 4.04AAVPTGDD172 pKa = 3.57AADD175 pKa = 4.21MYY177 pKa = 11.75AHH179 pKa = 6.43LQEE182 pKa = 4.6FAILQQMGMQEE193 pKa = 3.97MTYY196 pKa = 10.42EE197 pKa = 4.31KK198 pKa = 10.65YY199 pKa = 10.92LEE201 pKa = 4.34LFGGRR206 pKa = 11.84AAAAGQGKK214 pKa = 9.13PEE216 pKa = 4.38LLLYY220 pKa = 9.42DD221 pKa = 3.99QRR223 pKa = 11.84WKK225 pKa = 11.06KK226 pKa = 9.02PANTINPADD235 pKa = 4.1GTPSSAWVWNEE246 pKa = 3.45EE247 pKa = 4.17TKK249 pKa = 10.9LGSKK253 pKa = 9.82NKK255 pKa = 9.93RR256 pKa = 11.84FNEE259 pKa = 4.08PGFVIAFQTIRR270 pKa = 11.84PKK272 pKa = 10.15MFNGAVRR279 pKa = 11.84TSLVGNMWGFKK290 pKa = 9.68DD291 pKa = 3.78WYY293 pKa = 9.35PAYY296 pKa = 9.68TLDD299 pKa = 5.74DD300 pKa = 3.51PTATVKK306 pKa = 10.07TIANNDD312 pKa = 3.65PVFEE316 pKa = 4.3ASAYY320 pKa = 5.27TTGGTEE326 pKa = 5.02LIYY329 pKa = 10.97DD330 pKa = 4.11HH331 pKa = 7.49SDD333 pKa = 2.99LLAQGEE339 pKa = 4.35QFINDD344 pKa = 3.67YY345 pKa = 10.4APPYY349 pKa = 10.15ALPLSSGMVVEE360 pKa = 5.19GGGTPEE366 pKa = 4.06SFRR369 pKa = 11.84GEE371 pKa = 4.44YY372 pKa = 8.24CTSADD377 pKa = 3.69VDD379 pKa = 4.08ALFQDD384 pKa = 4.5GAKK387 pKa = 9.13TRR389 pKa = 11.84AFYY392 pKa = 10.96DD393 pKa = 4.79GIAFCNILGHH403 pKa = 6.09VADD406 pKa = 4.43NTPLKK411 pKa = 10.84
Molecular weight: 45.6 kDa
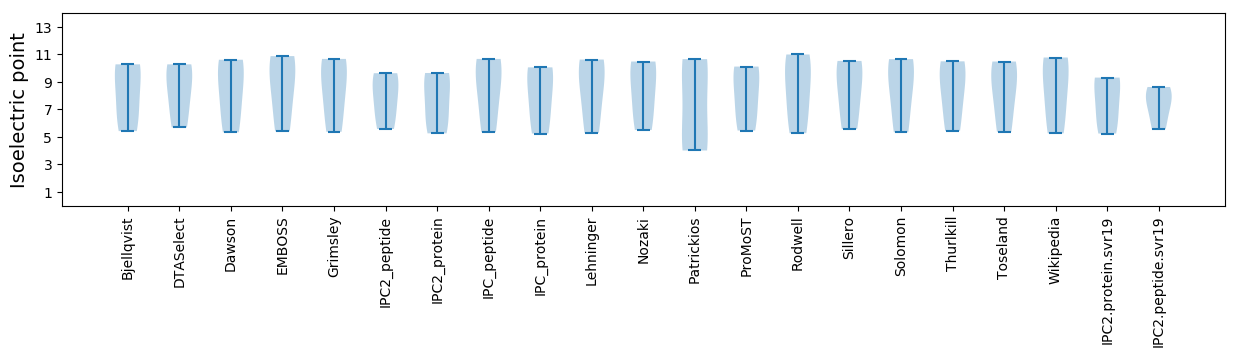
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z4QG15|A0A2Z4QG15_9VIRU Replication protein OS=Ruegeria phage vB_RpoMi-V15 OX=2218610 GN=vBRpoMiV15_02 PE=4 SV=1
MM1 pKa = 7.54EE2 pKa = 5.09FFRR5 pKa = 11.84YY6 pKa = 9.79SEE8 pKa = 4.29FQCKK12 pKa = 9.46CGCGMLPQPFVMDD25 pKa = 3.79NLDD28 pKa = 3.87HH29 pKa = 6.79IRR31 pKa = 11.84RR32 pKa = 11.84ALRR35 pKa = 11.84VPLVIHH41 pKa = 6.46SGARR45 pKa = 11.84CARR48 pKa = 11.84HH49 pKa = 5.09NARR52 pKa = 11.84VGGALLSQHH61 pKa = 5.21VKK63 pKa = 10.27GRR65 pKa = 11.84AVDD68 pKa = 3.48IRR70 pKa = 11.84WPEE73 pKa = 3.81GHH75 pKa = 6.35QSALLGLLQRR85 pKa = 11.84NGVRR89 pKa = 11.84GIGIYY94 pKa = 8.56PTFVHH99 pKa = 6.87ADD101 pKa = 3.01WRR103 pKa = 11.84EE104 pKa = 3.9GPSAFWVRR112 pKa = 11.84PEE114 pKa = 3.98GARR117 pKa = 11.84TT118 pKa = 3.29
MM1 pKa = 7.54EE2 pKa = 5.09FFRR5 pKa = 11.84YY6 pKa = 9.79SEE8 pKa = 4.29FQCKK12 pKa = 9.46CGCGMLPQPFVMDD25 pKa = 3.79NLDD28 pKa = 3.87HH29 pKa = 6.79IRR31 pKa = 11.84RR32 pKa = 11.84ALRR35 pKa = 11.84VPLVIHH41 pKa = 6.46SGARR45 pKa = 11.84CARR48 pKa = 11.84HH49 pKa = 5.09NARR52 pKa = 11.84VGGALLSQHH61 pKa = 5.21VKK63 pKa = 10.27GRR65 pKa = 11.84AVDD68 pKa = 3.48IRR70 pKa = 11.84WPEE73 pKa = 3.81GHH75 pKa = 6.35QSALLGLLQRR85 pKa = 11.84NGVRR89 pKa = 11.84GIGIYY94 pKa = 8.56PTFVHH99 pKa = 6.87ADD101 pKa = 3.01WRR103 pKa = 11.84EE104 pKa = 3.9GPSAFWVRR112 pKa = 11.84PEE114 pKa = 3.98GARR117 pKa = 11.84TT118 pKa = 3.29
Molecular weight: 13.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1294 |
118 |
451 |
323.5 |
35.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.505 ± 0.618 | 1.236 ± 0.475 |
5.255 ± 0.614 | 4.714 ± 0.074 |
3.941 ± 0.752 | 9.505 ± 0.976 |
3.246 ± 0.398 | 4.019 ± 0.492 |
5.873 ± 0.991 | 7.651 ± 0.346 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.318 ± 0.332 | 3.709 ± 0.669 |
6.646 ± 0.502 | 3.246 ± 0.442 |
7.419 ± 1.756 | 5.1 ± 0.436 |
5.41 ± 0.735 | 5.719 ± 0.331 |
2.087 ± 0.377 | 3.4 ± 0.353 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
