
Muribaculaceae bacterium Isolate-037 (Harlan)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Muribaculaceae; unclassified Muribaculaceae
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
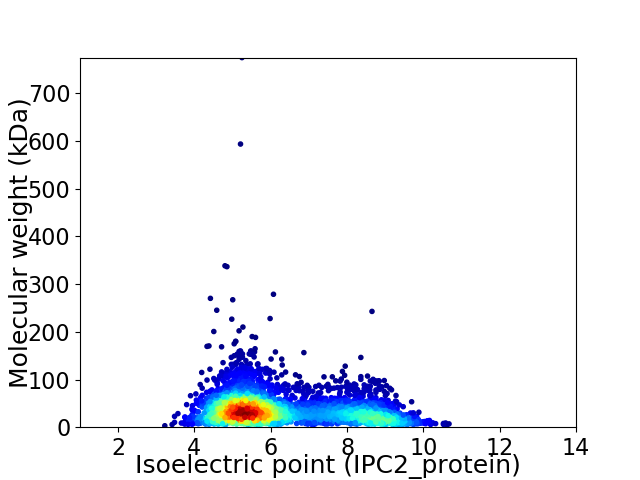
Virtual 2D-PAGE plot for 3984 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N2MFS3|A0A3N2MFS3_9BACT Uncharacterized protein OS=Muribaculaceae bacterium Isolate-037 (Harlan) OX=2486461 GN=EEL33_04705 PE=4 SV=1
MM1 pKa = 7.1SAAVSMAAPPQTGFYY16 pKa = 10.58LSGFNGEE23 pKa = 4.36SEE25 pKa = 4.16ATEE28 pKa = 4.46SNRR31 pKa = 11.84LTYY34 pKa = 10.72VPGNEE39 pKa = 3.85EE40 pKa = 4.71DD41 pKa = 3.83EE42 pKa = 5.04DD43 pKa = 3.7EE44 pKa = 5.09GIYY47 pKa = 10.61RR48 pKa = 11.84FINDD52 pKa = 3.14NFVISEE58 pKa = 4.16CQNGFFAIGEE68 pKa = 4.27EE69 pKa = 4.38GLKK72 pKa = 10.86LGFNVDD78 pKa = 3.11NDD80 pKa = 3.97FGFSNMVNQYY90 pKa = 9.23ATMTSLAEE98 pKa = 4.01NGPAINCEE106 pKa = 4.2LPAGTYY112 pKa = 10.06KK113 pKa = 10.91VILASMQEE121 pKa = 4.21EE122 pKa = 4.89GDD124 pKa = 3.87PMSWTMMFQSLNNDD138 pKa = 3.71DD139 pKa = 4.41EE140 pKa = 5.78AISYY144 pKa = 9.04YY145 pKa = 10.85LIGFNGNDD153 pKa = 4.02EE154 pKa = 4.47LSQDD158 pKa = 3.86NIFVAEE164 pKa = 4.09EE165 pKa = 4.31SEE167 pKa = 4.44DD168 pKa = 3.8GVIYY172 pKa = 8.53TYY174 pKa = 11.01PKK176 pKa = 10.54FLVEE180 pKa = 4.25DD181 pKa = 3.99CADD184 pKa = 3.54GFFVANSKK192 pKa = 11.24DD193 pKa = 3.66DD194 pKa = 3.93TLLGAAARR202 pKa = 11.84SDD204 pKa = 3.24KK205 pKa = 10.51VTDD208 pKa = 3.75EE209 pKa = 4.47NPFASLEE216 pKa = 4.1NSGDD220 pKa = 3.72AVQSSLTPGYY230 pKa = 8.77YY231 pKa = 8.94TVNFISMNGLNMISFLRR248 pKa = 11.84CEE250 pKa = 4.5DD251 pKa = 3.26QTPADD256 pKa = 3.64EE257 pKa = 4.3SVYY260 pKa = 11.14YY261 pKa = 10.98LLGFNGIEE269 pKa = 4.06NPTDD273 pKa = 3.12GVKK276 pKa = 10.43FVRR279 pKa = 11.84SVEE282 pKa = 3.94EE283 pKa = 3.56SSYY286 pKa = 11.16EE287 pKa = 3.95DD288 pKa = 3.96EE289 pKa = 4.7EE290 pKa = 4.37TGEE293 pKa = 4.46MISEE297 pKa = 4.16TVITYY302 pKa = 7.36TLKK305 pKa = 10.82NFDD308 pKa = 4.12IKK310 pKa = 11.07SCSEE314 pKa = 4.01DD315 pKa = 3.24GFIVTTEE322 pKa = 4.37DD323 pKa = 3.11GGFSYY328 pKa = 10.93GLSTMFVPMLGNTISLDD345 pKa = 3.71SPFGMMGIYY354 pKa = 9.85GKK356 pKa = 9.23PYY358 pKa = 10.3KK359 pKa = 10.32SALPEE364 pKa = 3.88GKK366 pKa = 10.18YY367 pKa = 10.57DD368 pKa = 3.89INFSTSDD375 pKa = 3.23TSASISFLPSEE386 pKa = 4.45DD387 pKa = 3.85SGVEE391 pKa = 4.18SIMTGDD397 pKa = 3.3NSEE400 pKa = 3.93AVYY403 pKa = 11.16YY404 pKa = 9.94DD405 pKa = 3.35LQGRR409 pKa = 11.84RR410 pKa = 11.84MINPDD415 pKa = 2.93KK416 pKa = 10.93GIYY419 pKa = 9.81ILVKK423 pKa = 9.81NGEE426 pKa = 3.81VSKK429 pKa = 10.69IVKK432 pKa = 9.63
MM1 pKa = 7.1SAAVSMAAPPQTGFYY16 pKa = 10.58LSGFNGEE23 pKa = 4.36SEE25 pKa = 4.16ATEE28 pKa = 4.46SNRR31 pKa = 11.84LTYY34 pKa = 10.72VPGNEE39 pKa = 3.85EE40 pKa = 4.71DD41 pKa = 3.83EE42 pKa = 5.04DD43 pKa = 3.7EE44 pKa = 5.09GIYY47 pKa = 10.61RR48 pKa = 11.84FINDD52 pKa = 3.14NFVISEE58 pKa = 4.16CQNGFFAIGEE68 pKa = 4.27EE69 pKa = 4.38GLKK72 pKa = 10.86LGFNVDD78 pKa = 3.11NDD80 pKa = 3.97FGFSNMVNQYY90 pKa = 9.23ATMTSLAEE98 pKa = 4.01NGPAINCEE106 pKa = 4.2LPAGTYY112 pKa = 10.06KK113 pKa = 10.91VILASMQEE121 pKa = 4.21EE122 pKa = 4.89GDD124 pKa = 3.87PMSWTMMFQSLNNDD138 pKa = 3.71DD139 pKa = 4.41EE140 pKa = 5.78AISYY144 pKa = 9.04YY145 pKa = 10.85LIGFNGNDD153 pKa = 4.02EE154 pKa = 4.47LSQDD158 pKa = 3.86NIFVAEE164 pKa = 4.09EE165 pKa = 4.31SEE167 pKa = 4.44DD168 pKa = 3.8GVIYY172 pKa = 8.53TYY174 pKa = 11.01PKK176 pKa = 10.54FLVEE180 pKa = 4.25DD181 pKa = 3.99CADD184 pKa = 3.54GFFVANSKK192 pKa = 11.24DD193 pKa = 3.66DD194 pKa = 3.93TLLGAAARR202 pKa = 11.84SDD204 pKa = 3.24KK205 pKa = 10.51VTDD208 pKa = 3.75EE209 pKa = 4.47NPFASLEE216 pKa = 4.1NSGDD220 pKa = 3.72AVQSSLTPGYY230 pKa = 8.77YY231 pKa = 8.94TVNFISMNGLNMISFLRR248 pKa = 11.84CEE250 pKa = 4.5DD251 pKa = 3.26QTPADD256 pKa = 3.64EE257 pKa = 4.3SVYY260 pKa = 11.14YY261 pKa = 10.98LLGFNGIEE269 pKa = 4.06NPTDD273 pKa = 3.12GVKK276 pKa = 10.43FVRR279 pKa = 11.84SVEE282 pKa = 3.94EE283 pKa = 3.56SSYY286 pKa = 11.16EE287 pKa = 3.95DD288 pKa = 3.96EE289 pKa = 4.7EE290 pKa = 4.37TGEE293 pKa = 4.46MISEE297 pKa = 4.16TVITYY302 pKa = 7.36TLKK305 pKa = 10.82NFDD308 pKa = 4.12IKK310 pKa = 11.07SCSEE314 pKa = 4.01DD315 pKa = 3.24GFIVTTEE322 pKa = 4.37DD323 pKa = 3.11GGFSYY328 pKa = 10.93GLSTMFVPMLGNTISLDD345 pKa = 3.71SPFGMMGIYY354 pKa = 9.85GKK356 pKa = 9.23PYY358 pKa = 10.3KK359 pKa = 10.32SALPEE364 pKa = 3.88GKK366 pKa = 10.18YY367 pKa = 10.57DD368 pKa = 3.89INFSTSDD375 pKa = 3.23TSASISFLPSEE386 pKa = 4.45DD387 pKa = 3.85SGVEE391 pKa = 4.18SIMTGDD397 pKa = 3.3NSEE400 pKa = 3.93AVYY403 pKa = 11.16YY404 pKa = 9.94DD405 pKa = 3.35LQGRR409 pKa = 11.84RR410 pKa = 11.84MINPDD415 pKa = 2.93KK416 pKa = 10.93GIYY419 pKa = 9.81ILVKK423 pKa = 9.81NGEE426 pKa = 3.81VSKK429 pKa = 10.69IVKK432 pKa = 9.63
Molecular weight: 47.36 kDa
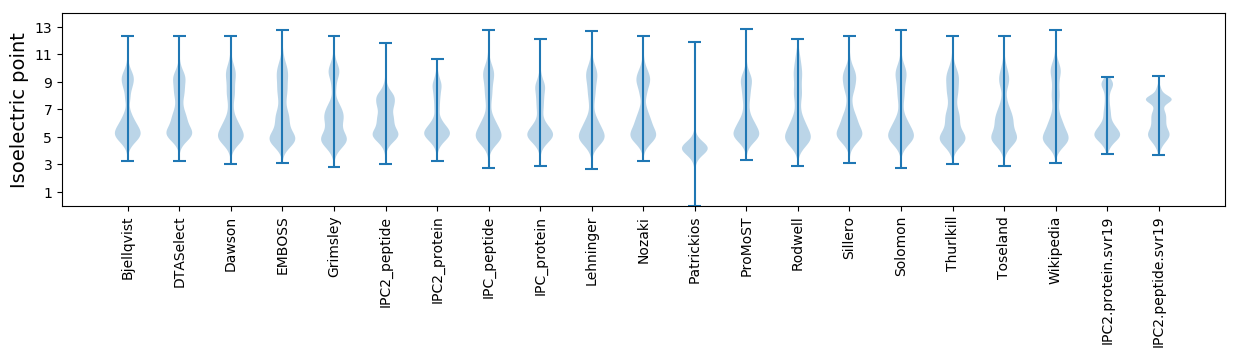
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N2M8V4|A0A3N2M8V4_9BACT DUF4099 domain-containing protein OS=Muribaculaceae bacterium Isolate-037 (Harlan) OX=2486461 GN=EEL33_09520 PE=4 SV=1
MM1 pKa = 7.43NGLEE5 pKa = 4.39KK6 pKa = 10.56ALQTLHH12 pKa = 7.03GYY14 pKa = 9.49GSRR17 pKa = 11.84HH18 pKa = 4.92SAIDD22 pKa = 3.05TGIQLFVTLGFIVAVVSVPTVDD44 pKa = 3.51PARR47 pKa = 11.84LIWFFAFPVIMAEE60 pKa = 4.03ALRR63 pKa = 11.84IGYY66 pKa = 9.16AHH68 pKa = 6.41VFCRR72 pKa = 11.84SLWLLPFIILIGIFNPIIDD91 pKa = 4.6RR92 pKa = 11.84QPAWEE97 pKa = 4.5IGNIVISKK105 pKa = 9.42GWISFISVVLRR116 pKa = 11.84GLLSVQALLILVDD129 pKa = 3.56SCGFNNICRR138 pKa = 11.84VLHH141 pKa = 5.47GVGVPSVLVTQLQMLYY157 pKa = 10.57RR158 pKa = 11.84FMGVLMEE165 pKa = 4.73EE166 pKa = 3.97ALIMHH171 pKa = 7.97RR172 pKa = 11.84ARR174 pKa = 11.84QSRR177 pKa = 11.84GFGRR181 pKa = 11.84KK182 pKa = 8.98SYY184 pKa = 9.14PLKK187 pKa = 10.07MWATFAGQLLMRR199 pKa = 11.84SLDD202 pKa = 3.29RR203 pKa = 11.84ASRR206 pKa = 11.84INRR209 pKa = 11.84AMLARR214 pKa = 11.84VYY216 pKa = 10.47DD217 pKa = 3.58SKK219 pKa = 11.04RR220 pKa = 11.84IHH222 pKa = 6.1KK223 pKa = 10.27AKK225 pKa = 10.26RR226 pKa = 11.84PKK228 pKa = 9.31LTRR231 pKa = 11.84NDD233 pKa = 4.01WIYY236 pKa = 10.81LAAWTTIILFIRR248 pKa = 11.84FIDD251 pKa = 3.56ISAIIDD257 pKa = 3.35KK258 pKa = 9.85WLSTHH263 pKa = 6.23LTT265 pKa = 3.57
MM1 pKa = 7.43NGLEE5 pKa = 4.39KK6 pKa = 10.56ALQTLHH12 pKa = 7.03GYY14 pKa = 9.49GSRR17 pKa = 11.84HH18 pKa = 4.92SAIDD22 pKa = 3.05TGIQLFVTLGFIVAVVSVPTVDD44 pKa = 3.51PARR47 pKa = 11.84LIWFFAFPVIMAEE60 pKa = 4.03ALRR63 pKa = 11.84IGYY66 pKa = 9.16AHH68 pKa = 6.41VFCRR72 pKa = 11.84SLWLLPFIILIGIFNPIIDD91 pKa = 4.6RR92 pKa = 11.84QPAWEE97 pKa = 4.5IGNIVISKK105 pKa = 9.42GWISFISVVLRR116 pKa = 11.84GLLSVQALLILVDD129 pKa = 3.56SCGFNNICRR138 pKa = 11.84VLHH141 pKa = 5.47GVGVPSVLVTQLQMLYY157 pKa = 10.57RR158 pKa = 11.84FMGVLMEE165 pKa = 4.73EE166 pKa = 3.97ALIMHH171 pKa = 7.97RR172 pKa = 11.84ARR174 pKa = 11.84QSRR177 pKa = 11.84GFGRR181 pKa = 11.84KK182 pKa = 8.98SYY184 pKa = 9.14PLKK187 pKa = 10.07MWATFAGQLLMRR199 pKa = 11.84SLDD202 pKa = 3.29RR203 pKa = 11.84ASRR206 pKa = 11.84INRR209 pKa = 11.84AMLARR214 pKa = 11.84VYY216 pKa = 10.47DD217 pKa = 3.58SKK219 pKa = 11.04RR220 pKa = 11.84IHH222 pKa = 6.1KK223 pKa = 10.27AKK225 pKa = 10.26RR226 pKa = 11.84PKK228 pKa = 9.31LTRR231 pKa = 11.84NDD233 pKa = 4.01WIYY236 pKa = 10.81LAAWTTIILFIRR248 pKa = 11.84FIDD251 pKa = 3.56ISAIIDD257 pKa = 3.35KK258 pKa = 9.85WLSTHH263 pKa = 6.23LTT265 pKa = 3.57
Molecular weight: 30.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1419065 |
29 |
6922 |
356.2 |
40.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.284 ± 0.047 | 1.202 ± 0.013 |
6.254 ± 0.028 | 6.369 ± 0.041 |
4.345 ± 0.027 | 6.96 ± 0.04 |
1.795 ± 0.017 | 6.962 ± 0.041 |
6.121 ± 0.034 | 8.599 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.812 ± 0.019 | 5.2 ± 0.032 |
3.934 ± 0.022 | 2.883 ± 0.023 |
5.106 ± 0.036 | 6.749 ± 0.033 |
5.499 ± 0.028 | 6.402 ± 0.032 |
1.318 ± 0.016 | 4.207 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
