
BD1-7 clade bacterium
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Spongiibacteraceae; unclassified Spongiibacteraceae; BD1-7 clade
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
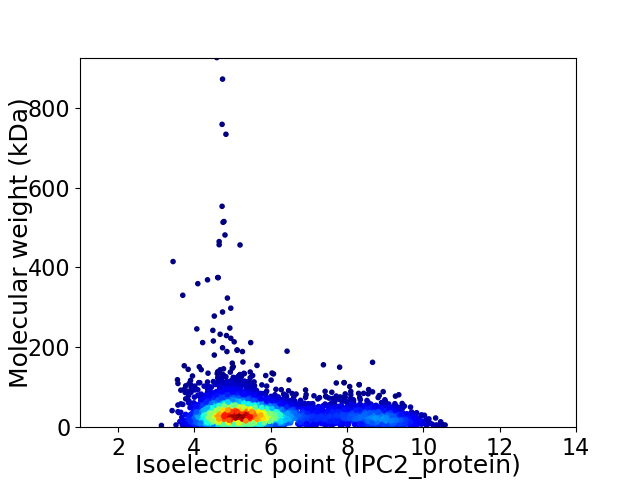
Virtual 2D-PAGE plot for 4504 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5S9PSZ9|A0A5S9PSZ9_9GAMM Uncharacterized protein OS=BD1-7 clade bacterium OX=2029982 GN=OPDIPICF_01203 PE=4 SV=1
MM1 pKa = 7.39TEE3 pKa = 4.17TMVHH7 pKa = 6.17PQQHH11 pKa = 6.07SKK13 pKa = 10.9CSAAMTNGIMQLSGWFASAMLVMFVCLIVSAAPAAFALTPANTLISNQATASYY66 pKa = 9.85KK67 pKa = 10.82SSDD70 pKa = 3.2GAQRR74 pKa = 11.84LATSNSVRR82 pKa = 11.84TIVTSVAAATLTQDD96 pKa = 3.04QTRR99 pKa = 11.84DD100 pKa = 3.41VLASGSVVFSHH111 pKa = 7.53IISNNGNVGDD121 pKa = 4.71SYY123 pKa = 11.46TITLTNQTGDD133 pKa = 3.71DD134 pKa = 4.1FNFTSLSAFADD145 pKa = 3.47ADD147 pKa = 4.05ANGQPDD153 pKa = 4.08TVQPLTEE160 pKa = 4.21TPVLAMQEE168 pKa = 4.12SFNIVIVGSIGTGEE182 pKa = 4.1NPGNEE187 pKa = 3.92AQVQVTATSILDD199 pKa = 3.46NSVVLNNTDD208 pKa = 3.2TAIVVDD214 pKa = 4.49DD215 pKa = 4.35AVVDD219 pKa = 3.74ITQSIDD225 pKa = 3.21LTQGIAGQTPIRR237 pKa = 11.84ITYY240 pKa = 8.58FYY242 pKa = 10.68VNSGSRR248 pKa = 11.84AATAITLGNLLPDD261 pKa = 3.73GFVYY265 pKa = 10.34QGNAVWSFGNLPLTDD280 pKa = 4.4ANEE283 pKa = 4.22EE284 pKa = 4.46TQGSAPSIIYY294 pKa = 9.72CAYY297 pKa = 10.37QPSCTQADD305 pKa = 3.79RR306 pKa = 11.84LSFVIDD312 pKa = 3.59EE313 pKa = 4.81LGAGQSGSVSFDD325 pKa = 2.84IAIDD329 pKa = 3.62ATQLPGVIQNTATYY343 pKa = 10.3DD344 pKa = 3.59YY345 pKa = 10.97LPFNGTQITGRR356 pKa = 11.84STNTVPFTVVGDD368 pKa = 3.92EE369 pKa = 4.5SLTLTDD375 pKa = 5.53DD376 pKa = 3.84SVTDD380 pKa = 4.01AVPGTTVFVTHH391 pKa = 6.65TVTNTGTGNDD401 pKa = 3.48TFNIRR406 pKa = 11.84VNQISSNFPVGTVFFLYY423 pKa = 10.48QSDD426 pKa = 3.96GLTPLNDD433 pKa = 3.76SNLDD437 pKa = 3.36GDD439 pKa = 5.55LDD441 pKa = 4.06TGPLPKK447 pKa = 10.34DD448 pKa = 3.41DD449 pKa = 5.03DD450 pKa = 4.39VNIIIGITLPPSAATGTYY468 pKa = 10.18SVGLRR473 pKa = 11.84GTSLQSNNTFADD485 pKa = 4.94AIATVSVLAPLLAVDD500 pKa = 4.49LTGIAANGDD509 pKa = 4.0SACDD513 pKa = 3.64ADD515 pKa = 4.84ADD517 pKa = 4.06NCGFEE522 pKa = 5.02AFDD525 pKa = 4.11PADD528 pKa = 3.73PAVISQTIDD537 pKa = 2.94PGQTANFVLYY547 pKa = 10.52ARR549 pKa = 11.84NLSEE553 pKa = 5.46IIDD556 pKa = 3.91LFGFSASTDD565 pKa = 3.37NTFGSQTLPAGWSISYY581 pKa = 9.41TEE583 pKa = 4.46LTDD586 pKa = 3.74EE587 pKa = 5.03VIASTGTLSPGGVKK601 pKa = 10.2AFKK604 pKa = 10.71VAVTTSEE611 pKa = 4.02TALATTEE618 pKa = 3.95DD619 pKa = 4.72LYY621 pKa = 10.97IQVLSATSLRR631 pKa = 11.84SDD633 pKa = 3.74RR634 pKa = 11.84LRR636 pKa = 11.84LSISINEE643 pKa = 4.18VQSVSLGPDD652 pKa = 2.99NASTEE657 pKa = 4.18EE658 pKa = 3.96PNQFVIYY665 pKa = 10.12RR666 pKa = 11.84HH667 pKa = 5.51EE668 pKa = 4.14LVNDD672 pKa = 3.88GNAAVTFDD680 pKa = 4.71GIAVALTTPDD690 pKa = 2.87WSIALYY696 pKa = 10.43EE697 pKa = 4.16DD698 pKa = 3.99TAGGTVDD705 pKa = 3.67EE706 pKa = 5.14WDD708 pKa = 3.88VDD710 pKa = 3.78DD711 pKa = 5.89QIINGSFSVAAFSSKK726 pKa = 9.07VVYY729 pKa = 10.33VRR731 pKa = 11.84AFIPSSATPGQVEE744 pKa = 4.52TATVTATFNAGVDD757 pKa = 3.51QVSAIDD763 pKa = 4.25VITANPFDD771 pKa = 4.02IDD773 pKa = 3.62IVKK776 pKa = 10.36AQAKK780 pKa = 9.22DD781 pKa = 3.66ANCDD785 pKa = 3.31GTEE788 pKa = 4.01DD789 pKa = 3.4SAFAVGSFGALPGEE803 pKa = 4.58CIVYY807 pKa = 10.37RR808 pKa = 11.84LRR810 pKa = 11.84VSNFSDD816 pKa = 2.82ITAVNAVIRR825 pKa = 11.84DD826 pKa = 3.74ATPEE830 pKa = 3.92YY831 pKa = 9.75TFYY834 pKa = 11.24SATQPATRR842 pKa = 11.84CTPAVCVFVSEE853 pKa = 4.62PAASQTGEE861 pKa = 4.03LEE863 pKa = 4.25INVGDD868 pKa = 3.67IDD870 pKa = 4.66AGNTVDD876 pKa = 6.01FFFSVKK882 pKa = 10.33VSDD885 pKa = 3.73TT886 pKa = 3.44
MM1 pKa = 7.39TEE3 pKa = 4.17TMVHH7 pKa = 6.17PQQHH11 pKa = 6.07SKK13 pKa = 10.9CSAAMTNGIMQLSGWFASAMLVMFVCLIVSAAPAAFALTPANTLISNQATASYY66 pKa = 9.85KK67 pKa = 10.82SSDD70 pKa = 3.2GAQRR74 pKa = 11.84LATSNSVRR82 pKa = 11.84TIVTSVAAATLTQDD96 pKa = 3.04QTRR99 pKa = 11.84DD100 pKa = 3.41VLASGSVVFSHH111 pKa = 7.53IISNNGNVGDD121 pKa = 4.71SYY123 pKa = 11.46TITLTNQTGDD133 pKa = 3.71DD134 pKa = 4.1FNFTSLSAFADD145 pKa = 3.47ADD147 pKa = 4.05ANGQPDD153 pKa = 4.08TVQPLTEE160 pKa = 4.21TPVLAMQEE168 pKa = 4.12SFNIVIVGSIGTGEE182 pKa = 4.1NPGNEE187 pKa = 3.92AQVQVTATSILDD199 pKa = 3.46NSVVLNNTDD208 pKa = 3.2TAIVVDD214 pKa = 4.49DD215 pKa = 4.35AVVDD219 pKa = 3.74ITQSIDD225 pKa = 3.21LTQGIAGQTPIRR237 pKa = 11.84ITYY240 pKa = 8.58FYY242 pKa = 10.68VNSGSRR248 pKa = 11.84AATAITLGNLLPDD261 pKa = 3.73GFVYY265 pKa = 10.34QGNAVWSFGNLPLTDD280 pKa = 4.4ANEE283 pKa = 4.22EE284 pKa = 4.46TQGSAPSIIYY294 pKa = 9.72CAYY297 pKa = 10.37QPSCTQADD305 pKa = 3.79RR306 pKa = 11.84LSFVIDD312 pKa = 3.59EE313 pKa = 4.81LGAGQSGSVSFDD325 pKa = 2.84IAIDD329 pKa = 3.62ATQLPGVIQNTATYY343 pKa = 10.3DD344 pKa = 3.59YY345 pKa = 10.97LPFNGTQITGRR356 pKa = 11.84STNTVPFTVVGDD368 pKa = 3.92EE369 pKa = 4.5SLTLTDD375 pKa = 5.53DD376 pKa = 3.84SVTDD380 pKa = 4.01AVPGTTVFVTHH391 pKa = 6.65TVTNTGTGNDD401 pKa = 3.48TFNIRR406 pKa = 11.84VNQISSNFPVGTVFFLYY423 pKa = 10.48QSDD426 pKa = 3.96GLTPLNDD433 pKa = 3.76SNLDD437 pKa = 3.36GDD439 pKa = 5.55LDD441 pKa = 4.06TGPLPKK447 pKa = 10.34DD448 pKa = 3.41DD449 pKa = 5.03DD450 pKa = 4.39VNIIIGITLPPSAATGTYY468 pKa = 10.18SVGLRR473 pKa = 11.84GTSLQSNNTFADD485 pKa = 4.94AIATVSVLAPLLAVDD500 pKa = 4.49LTGIAANGDD509 pKa = 4.0SACDD513 pKa = 3.64ADD515 pKa = 4.84ADD517 pKa = 4.06NCGFEE522 pKa = 5.02AFDD525 pKa = 4.11PADD528 pKa = 3.73PAVISQTIDD537 pKa = 2.94PGQTANFVLYY547 pKa = 10.52ARR549 pKa = 11.84NLSEE553 pKa = 5.46IIDD556 pKa = 3.91LFGFSASTDD565 pKa = 3.37NTFGSQTLPAGWSISYY581 pKa = 9.41TEE583 pKa = 4.46LTDD586 pKa = 3.74EE587 pKa = 5.03VIASTGTLSPGGVKK601 pKa = 10.2AFKK604 pKa = 10.71VAVTTSEE611 pKa = 4.02TALATTEE618 pKa = 3.95DD619 pKa = 4.72LYY621 pKa = 10.97IQVLSATSLRR631 pKa = 11.84SDD633 pKa = 3.74RR634 pKa = 11.84LRR636 pKa = 11.84LSISINEE643 pKa = 4.18VQSVSLGPDD652 pKa = 2.99NASTEE657 pKa = 4.18EE658 pKa = 3.96PNQFVIYY665 pKa = 10.12RR666 pKa = 11.84HH667 pKa = 5.51EE668 pKa = 4.14LVNDD672 pKa = 3.88GNAAVTFDD680 pKa = 4.71GIAVALTTPDD690 pKa = 2.87WSIALYY696 pKa = 10.43EE697 pKa = 4.16DD698 pKa = 3.99TAGGTVDD705 pKa = 3.67EE706 pKa = 5.14WDD708 pKa = 3.88VDD710 pKa = 3.78DD711 pKa = 5.89QIINGSFSVAAFSSKK726 pKa = 9.07VVYY729 pKa = 10.33VRR731 pKa = 11.84AFIPSSATPGQVEE744 pKa = 4.52TATVTATFNAGVDD757 pKa = 3.51QVSAIDD763 pKa = 4.25VITANPFDD771 pKa = 4.02IDD773 pKa = 3.62IVKK776 pKa = 10.36AQAKK780 pKa = 9.22DD781 pKa = 3.66ANCDD785 pKa = 3.31GTEE788 pKa = 4.01DD789 pKa = 3.4SAFAVGSFGALPGEE803 pKa = 4.58CIVYY807 pKa = 10.37RR808 pKa = 11.84LRR810 pKa = 11.84VSNFSDD816 pKa = 2.82ITAVNAVIRR825 pKa = 11.84DD826 pKa = 3.74ATPEE830 pKa = 3.92YY831 pKa = 9.75TFYY834 pKa = 11.24SATQPATRR842 pKa = 11.84CTPAVCVFVSEE853 pKa = 4.62PAASQTGEE861 pKa = 4.03LEE863 pKa = 4.25INVGDD868 pKa = 3.67IDD870 pKa = 4.66AGNTVDD876 pKa = 6.01FFFSVKK882 pKa = 10.33VSDD885 pKa = 3.73TT886 pKa = 3.44
Molecular weight: 92.68 kDa
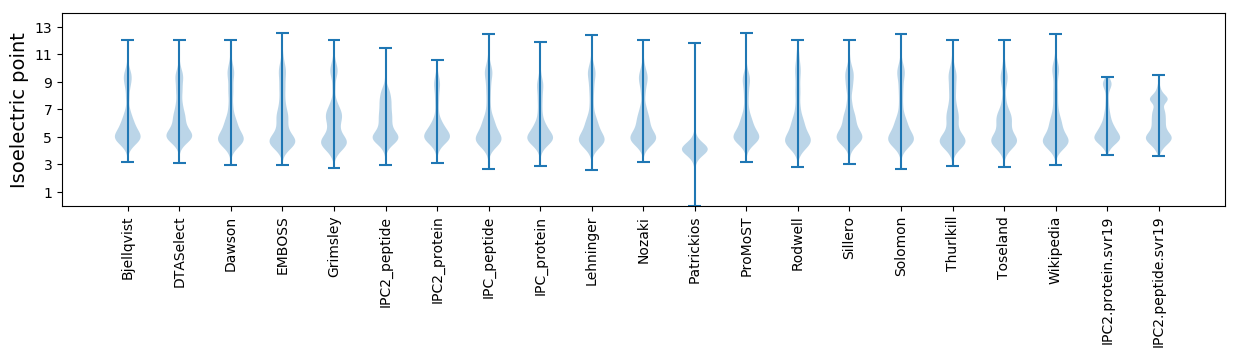
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5S9PUD2|A0A5S9PUD2_9GAMM Bacterioferritin OS=BD1-7 clade bacterium OX=2029982 GN=bfr PE=3 SV=1
MM1 pKa = 7.06FAAITHH7 pKa = 5.55QSRR10 pKa = 11.84HH11 pKa = 5.74SGHH14 pKa = 6.18SFVPASRR21 pKa = 11.84FLALNKK27 pKa = 9.64AARR30 pKa = 11.84VGTRR34 pKa = 11.84LNKK37 pKa = 9.97KK38 pKa = 8.61PALASMHH45 pKa = 6.32VSDD48 pKa = 5.48SYY50 pKa = 11.65CADD53 pKa = 3.29ASSRR57 pKa = 11.84RR58 pKa = 11.84CSGGLLRR65 pKa = 11.84LPPTVDD71 pKa = 2.94AFRR74 pKa = 11.84SEE76 pKa = 3.95PHH78 pKa = 5.38TLYY81 pKa = 10.18MSRR84 pKa = 11.84LMAMRR89 pKa = 11.84VKK91 pKa = 10.72ACGPSFLKK99 pKa = 10.15TDD101 pKa = 3.43SDD103 pKa = 4.02PPRR106 pKa = 11.84NGGSQRR112 pKa = 11.84VFTLPEE118 pKa = 3.97ILNSSS123 pKa = 3.6
MM1 pKa = 7.06FAAITHH7 pKa = 5.55QSRR10 pKa = 11.84HH11 pKa = 5.74SGHH14 pKa = 6.18SFVPASRR21 pKa = 11.84FLALNKK27 pKa = 9.64AARR30 pKa = 11.84VGTRR34 pKa = 11.84LNKK37 pKa = 9.97KK38 pKa = 8.61PALASMHH45 pKa = 6.32VSDD48 pKa = 5.48SYY50 pKa = 11.65CADD53 pKa = 3.29ASSRR57 pKa = 11.84RR58 pKa = 11.84CSGGLLRR65 pKa = 11.84LPPTVDD71 pKa = 2.94AFRR74 pKa = 11.84SEE76 pKa = 3.95PHH78 pKa = 5.38TLYY81 pKa = 10.18MSRR84 pKa = 11.84LMAMRR89 pKa = 11.84VKK91 pKa = 10.72ACGPSFLKK99 pKa = 10.15TDD101 pKa = 3.43SDD103 pKa = 4.02PPRR106 pKa = 11.84NGGSQRR112 pKa = 11.84VFTLPEE118 pKa = 3.97ILNSSS123 pKa = 3.6
Molecular weight: 13.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1561466 |
29 |
8590 |
346.7 |
38.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.393 ± 0.04 | 1.099 ± 0.016 |
6.332 ± 0.038 | 5.689 ± 0.04 |
4.087 ± 0.024 | 6.915 ± 0.037 |
2.369 ± 0.023 | 6.044 ± 0.03 |
4.364 ± 0.038 | 10.012 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.432 ± 0.029 | 4.127 ± 0.03 |
4.258 ± 0.027 | 4.418 ± 0.034 |
4.923 ± 0.031 | 6.66 ± 0.039 |
5.661 ± 0.043 | 6.923 ± 0.034 |
1.282 ± 0.016 | 2.996 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
