
Maize rayado fino virus (isolate Costa Rica/Guapiles) (MRFV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Tymoviridae; Marafivirus; Maize rayado fino virus
Average proteome isoelectric point is 7.38
Get precalculated fractions of proteins
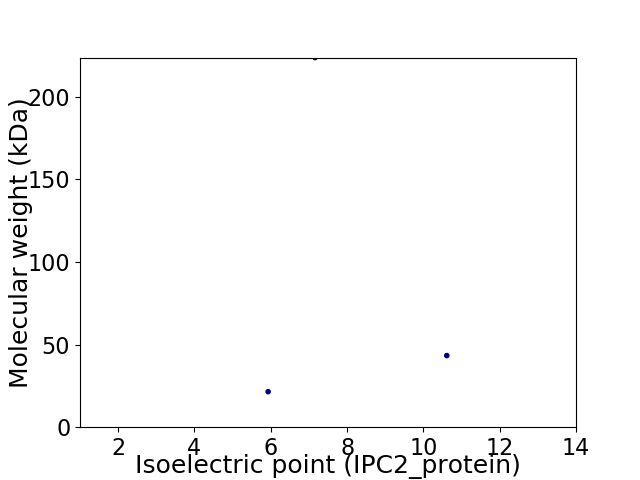
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q91TW9-2|POLG-2_MRFVC Isoform of Q91TW9 Isoform Subgenomic capsid protein CP2 of Genome polyprotein OS=Maize rayado fino virus (isolate Costa Rica/Guapiles) OX=652669 GN=ORF1 PE=4 SV=1
MM1 pKa = 7.98ADD3 pKa = 3.15NATQVGPVPPRR14 pKa = 11.84DD15 pKa = 3.61DD16 pKa = 3.59RR17 pKa = 11.84VDD19 pKa = 3.74RR20 pKa = 11.84QPPLPDD26 pKa = 3.57PPRR29 pKa = 11.84VLEE32 pKa = 4.02TAPSHH37 pKa = 7.1FLDD40 pKa = 6.1LPFQWKK46 pKa = 7.96VTDD49 pKa = 3.78FTGYY53 pKa = 10.43AAYY56 pKa = 10.18HH57 pKa = 5.47GTDD60 pKa = 3.96DD61 pKa = 4.23LVASAVLTTLCAPYY75 pKa = 10.22RR76 pKa = 11.84HH77 pKa = 6.64AEE79 pKa = 3.94LLYY82 pKa = 10.82VEE84 pKa = 5.48ISVAPCPPSFSKK96 pKa = 10.86PIMFTVVWTPATLSPRR112 pKa = 11.84DD113 pKa = 3.72GKK115 pKa = 9.3EE116 pKa = 3.21TDD118 pKa = 3.7YY119 pKa = 11.7YY120 pKa = 10.94GGRR123 pKa = 11.84QITVGGPVMLSSTTAVPADD142 pKa = 4.22LARR145 pKa = 11.84MNPFIKK151 pKa = 10.54SSVSYY156 pKa = 10.97NDD158 pKa = 3.26TPRR161 pKa = 11.84WTMSVPAVTGGDD173 pKa = 3.51TKK175 pKa = 10.95IPLATAFVRR184 pKa = 11.84GIVRR188 pKa = 11.84VRR190 pKa = 11.84APSGAATPSAA200 pKa = 3.93
MM1 pKa = 7.98ADD3 pKa = 3.15NATQVGPVPPRR14 pKa = 11.84DD15 pKa = 3.61DD16 pKa = 3.59RR17 pKa = 11.84VDD19 pKa = 3.74RR20 pKa = 11.84QPPLPDD26 pKa = 3.57PPRR29 pKa = 11.84VLEE32 pKa = 4.02TAPSHH37 pKa = 7.1FLDD40 pKa = 6.1LPFQWKK46 pKa = 7.96VTDD49 pKa = 3.78FTGYY53 pKa = 10.43AAYY56 pKa = 10.18HH57 pKa = 5.47GTDD60 pKa = 3.96DD61 pKa = 4.23LVASAVLTTLCAPYY75 pKa = 10.22RR76 pKa = 11.84HH77 pKa = 6.64AEE79 pKa = 3.94LLYY82 pKa = 10.82VEE84 pKa = 5.48ISVAPCPPSFSKK96 pKa = 10.86PIMFTVVWTPATLSPRR112 pKa = 11.84DD113 pKa = 3.72GKK115 pKa = 9.3EE116 pKa = 3.21TDD118 pKa = 3.7YY119 pKa = 11.7YY120 pKa = 10.94GGRR123 pKa = 11.84QITVGGPVMLSSTTAVPADD142 pKa = 4.22LARR145 pKa = 11.84MNPFIKK151 pKa = 10.54SSVSYY156 pKa = 10.97NDD158 pKa = 3.26TPRR161 pKa = 11.84WTMSVPAVTGGDD173 pKa = 3.51TKK175 pKa = 10.95IPLATAFVRR184 pKa = 11.84GIVRR188 pKa = 11.84VRR190 pKa = 11.84APSGAATPSAA200 pKa = 3.93
Molecular weight: 21.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q91TW9|POLG_MRFVC Genome polyprotein OS=Maize rayado fino virus (isolate Costa Rica/Guapiles) OX=652669 GN=ORF1 PE=4 SV=1
MM1 pKa = 8.0PLTPTPSIRR10 pKa = 11.84PSRR13 pKa = 11.84PTSFSMSGPTTLGVRR28 pKa = 11.84QTSCSSSLRR37 pKa = 11.84SSPSSSPDD45 pKa = 3.09SPTSPTSSTTGSCPKK60 pKa = 9.29TPPGTPPLPRR70 pKa = 11.84TSRR73 pKa = 11.84TARR76 pKa = 11.84PSSCMMLSCITPQGRR91 pKa = 11.84SLTSSSSVPSSRR103 pKa = 11.84RR104 pKa = 11.84SMPPLSFRR112 pKa = 11.84LSRR115 pKa = 11.84ASRR118 pKa = 11.84ISRR121 pKa = 11.84SIRR124 pKa = 11.84KK125 pKa = 7.44FTASVFRR132 pKa = 11.84VLTLSTSRR140 pKa = 11.84RR141 pKa = 11.84ATQPPTTRR149 pKa = 11.84SRR151 pKa = 11.84VRR153 pKa = 11.84PSTGSRR159 pKa = 11.84PPVSPLVTSSSPSPFSTLSARR180 pKa = 11.84SIPSSSSVAALPSFRR195 pKa = 11.84PKK197 pKa = 10.09TLLPSAFRR205 pKa = 11.84TPSPSPLPPPSTRR218 pKa = 11.84TFVTGWSPARR228 pKa = 11.84CTTRR232 pKa = 11.84SSIMSEE238 pKa = 3.86PSAFGLPTPPASSGLRR254 pKa = 11.84SASLSTAGSPLPPGTTFSTSRR275 pKa = 11.84SRR277 pKa = 11.84PPPFVPTPRR286 pKa = 11.84IPFSSRR292 pKa = 11.84PSLACPTGFAPTLGRR307 pKa = 11.84SGAWLPPRR315 pKa = 11.84PLSLPGPLPVPSAGSSPFTPTVSGCSASTSSAGGSGLVSHH355 pKa = 6.8SMAPSQGFFGRR366 pKa = 11.84PIQPAARR373 pKa = 11.84QCSLPTPPLSAKK385 pKa = 10.39SLLVWQTGVLPPPSGRR401 pKa = 11.84VCSPRR406 pKa = 11.84PLRR409 pKa = 11.84RR410 pKa = 11.84PGSPTQPWPP419 pKa = 3.27
MM1 pKa = 8.0PLTPTPSIRR10 pKa = 11.84PSRR13 pKa = 11.84PTSFSMSGPTTLGVRR28 pKa = 11.84QTSCSSSLRR37 pKa = 11.84SSPSSSPDD45 pKa = 3.09SPTSPTSSTTGSCPKK60 pKa = 9.29TPPGTPPLPRR70 pKa = 11.84TSRR73 pKa = 11.84TARR76 pKa = 11.84PSSCMMLSCITPQGRR91 pKa = 11.84SLTSSSSVPSSRR103 pKa = 11.84RR104 pKa = 11.84SMPPLSFRR112 pKa = 11.84LSRR115 pKa = 11.84ASRR118 pKa = 11.84ISRR121 pKa = 11.84SIRR124 pKa = 11.84KK125 pKa = 7.44FTASVFRR132 pKa = 11.84VLTLSTSRR140 pKa = 11.84RR141 pKa = 11.84ATQPPTTRR149 pKa = 11.84SRR151 pKa = 11.84VRR153 pKa = 11.84PSTGSRR159 pKa = 11.84PPVSPLVTSSSPSPFSTLSARR180 pKa = 11.84SIPSSSSVAALPSFRR195 pKa = 11.84PKK197 pKa = 10.09TLLPSAFRR205 pKa = 11.84TPSPSPLPPPSTRR218 pKa = 11.84TFVTGWSPARR228 pKa = 11.84CTTRR232 pKa = 11.84SSIMSEE238 pKa = 3.86PSAFGLPTPPASSGLRR254 pKa = 11.84SASLSTAGSPLPPGTTFSTSRR275 pKa = 11.84SRR277 pKa = 11.84PPPFVPTPRR286 pKa = 11.84IPFSSRR292 pKa = 11.84PSLACPTGFAPTLGRR307 pKa = 11.84SGAWLPPRR315 pKa = 11.84PLSLPGPLPVPSAGSSPFTPTVSGCSASTSSAGGSGLVSHH355 pKa = 6.8SMAPSQGFFGRR366 pKa = 11.84PIQPAARR373 pKa = 11.84QCSLPTPPLSAKK385 pKa = 10.39SLLVWQTGVLPPPSGRR401 pKa = 11.84VCSPRR406 pKa = 11.84PLRR409 pKa = 11.84RR410 pKa = 11.84PGSPTQPWPP419 pKa = 3.27
Molecular weight: 43.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2646 |
200 |
2027 |
882.0 |
96.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.788 ± 1.458 | 1.474 ± 0.206 |
4.875 ± 1.467 | 3.175 ± 1.185 |
4.724 ± 0.497 | 5.329 ± 0.319 |
2.91 ± 1.125 | 2.91 ± 0.347 |
2.192 ± 0.41 | 9.864 ± 1.208 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.55 ± 0.216 | 1.587 ± 0.574 |
11.489 ± 2.816 | 2.683 ± 0.367 |
7.596 ± 0.666 | 10.847 ± 3.935 |
6.916 ± 1.848 | 6.16 ± 0.777 |
1.512 ± 0.199 | 2.419 ± 0.767 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
