
Alkalispirochaeta americana
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Spirochaetaceae; Alkalispirochaeta
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
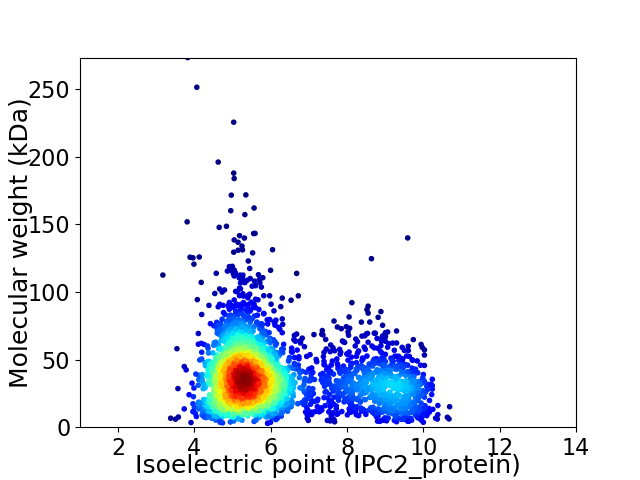
Virtual 2D-PAGE plot for 2891 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
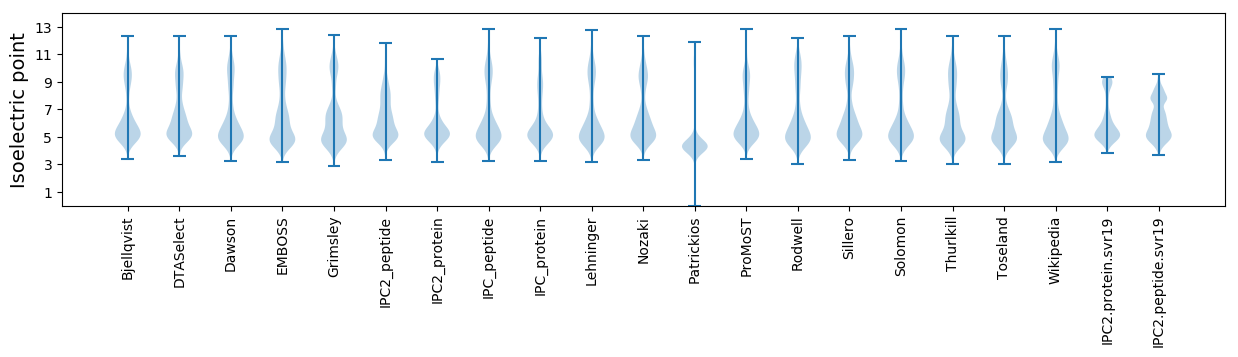
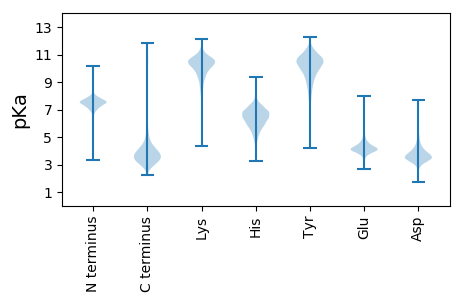
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1N6PQN8|A0A1N6PQN8_9SPIO Uncharacterized protein OS=Alkalispirochaeta americana OX=159291 GN=SAMN05920897_10386 PE=4 SV=1
MM1 pKa = 6.76KK2 pKa = 9.68TKK4 pKa = 10.54NALFSACPSLHH15 pKa = 6.41SSFFLGVAWFFLIFLALGGCYY36 pKa = 7.32WTPDD40 pKa = 3.3EE41 pKa = 4.51SRR43 pKa = 11.84GSISLRR49 pKa = 11.84IQAPSSGEE57 pKa = 3.76QVTASDD63 pKa = 4.11TVGEE67 pKa = 4.1PAYY70 pKa = 10.76LLLVYY75 pKa = 10.4VIEE78 pKa = 4.33EE79 pKa = 3.84AFIRR83 pKa = 11.84QDD85 pKa = 3.12PEE87 pKa = 4.01GNMEE91 pKa = 5.02HH92 pKa = 7.56IFGLWEE98 pKa = 3.88ASFDD102 pKa = 3.81EE103 pKa = 4.66EE104 pKa = 4.75DD105 pKa = 4.39FEE107 pKa = 5.6GADD110 pKa = 3.84SSPGSLGSISLGTSALASQFQGFAEE135 pKa = 4.04LLGYY139 pKa = 9.9EE140 pKa = 4.39SSAGGTILFSNLNPDD155 pKa = 2.63SSYY158 pKa = 11.23LVVAEE163 pKa = 4.0VHH165 pKa = 6.21SLEE168 pKa = 4.15EE169 pKa = 3.98LVGEE173 pKa = 4.24GQEE176 pKa = 3.84DD177 pKa = 3.2SRR179 pKa = 11.84YY180 pKa = 9.87SFRR183 pKa = 11.84TTTVRR188 pKa = 11.84SGEE191 pKa = 4.01NRR193 pKa = 11.84TVNLDD198 pKa = 3.24LDD200 pKa = 3.92KK201 pKa = 11.11TLNQYY206 pKa = 11.22LEE208 pKa = 4.31FLNARR213 pKa = 11.84YY214 pKa = 9.54GLSEE218 pKa = 4.9PDD220 pKa = 4.12DD221 pKa = 4.65PSDD224 pKa = 3.61GSIEE228 pKa = 3.8ITANLVNPEE237 pKa = 4.18DD238 pKa = 4.04EE239 pKa = 4.93EE240 pKa = 5.35IEE242 pKa = 4.17LTADD246 pKa = 3.31AEE248 pKa = 4.64VNLDD252 pKa = 4.27EE253 pKa = 5.67ISEE256 pKa = 4.35LTFTAPEE263 pKa = 5.09GYY265 pKa = 10.1DD266 pKa = 3.39SYY268 pKa = 11.78EE269 pKa = 3.66WTVNNMEE276 pKa = 4.34GWDD279 pKa = 3.78NSNTITISEE288 pKa = 4.36SEE290 pKa = 3.93ISDD293 pKa = 3.85VIFPGLNTVDD303 pKa = 4.97LEE305 pKa = 4.47VEE307 pKa = 4.12LGGEE311 pKa = 4.61VYY313 pKa = 10.64HH314 pKa = 6.34GTFTFTAIEE323 pKa = 4.46TILGEE328 pKa = 4.15LVIEE332 pKa = 5.19AILVNPEE339 pKa = 4.27DD340 pKa = 3.85EE341 pKa = 5.07QITLDD346 pKa = 4.15ADD348 pKa = 3.7AEE350 pKa = 4.57VNLDD354 pKa = 4.27EE355 pKa = 5.67ISEE358 pKa = 4.35LTFTAPEE365 pKa = 5.21GYY367 pKa = 10.18DD368 pKa = 3.41SYY370 pKa = 12.02NWTVNNMEE378 pKa = 4.57DD379 pKa = 3.16WGNSNTITISEE390 pKa = 4.17SDD392 pKa = 3.2ISDD395 pKa = 3.73VIFPGLNTVDD405 pKa = 4.97LEE407 pKa = 4.47VEE409 pKa = 4.12LGGEE413 pKa = 4.61VYY415 pKa = 10.64HH416 pKa = 6.34GTFTFTAIEE425 pKa = 4.46TILGEE430 pKa = 4.15LVIEE434 pKa = 5.19AILVNPEE441 pKa = 3.96DD442 pKa = 4.07EE443 pKa = 4.9EE444 pKa = 5.27IEE446 pKa = 4.16LTADD450 pKa = 3.3TEE452 pKa = 4.58VNIDD456 pKa = 4.43EE457 pKa = 4.32ITEE460 pKa = 4.19LTFTAPTGYY469 pKa = 10.44DD470 pKa = 3.17SYY472 pKa = 11.29EE473 pKa = 3.71WTVNDD478 pKa = 3.72MEE480 pKa = 4.9GWDD483 pKa = 3.83NSNTITIPEE492 pKa = 4.1SDD494 pKa = 3.27ISDD497 pKa = 3.99VIKK500 pKa = 10.84LGQNTVDD507 pKa = 4.13LVVEE511 pKa = 4.27KK512 pKa = 10.98AGALYY517 pKa = 9.69SHH519 pKa = 7.48RR520 pKa = 11.84FTFTAISDD528 pKa = 3.62
MM1 pKa = 6.76KK2 pKa = 9.68TKK4 pKa = 10.54NALFSACPSLHH15 pKa = 6.41SSFFLGVAWFFLIFLALGGCYY36 pKa = 7.32WTPDD40 pKa = 3.3EE41 pKa = 4.51SRR43 pKa = 11.84GSISLRR49 pKa = 11.84IQAPSSGEE57 pKa = 3.76QVTASDD63 pKa = 4.11TVGEE67 pKa = 4.1PAYY70 pKa = 10.76LLLVYY75 pKa = 10.4VIEE78 pKa = 4.33EE79 pKa = 3.84AFIRR83 pKa = 11.84QDD85 pKa = 3.12PEE87 pKa = 4.01GNMEE91 pKa = 5.02HH92 pKa = 7.56IFGLWEE98 pKa = 3.88ASFDD102 pKa = 3.81EE103 pKa = 4.66EE104 pKa = 4.75DD105 pKa = 4.39FEE107 pKa = 5.6GADD110 pKa = 3.84SSPGSLGSISLGTSALASQFQGFAEE135 pKa = 4.04LLGYY139 pKa = 9.9EE140 pKa = 4.39SSAGGTILFSNLNPDD155 pKa = 2.63SSYY158 pKa = 11.23LVVAEE163 pKa = 4.0VHH165 pKa = 6.21SLEE168 pKa = 4.15EE169 pKa = 3.98LVGEE173 pKa = 4.24GQEE176 pKa = 3.84DD177 pKa = 3.2SRR179 pKa = 11.84YY180 pKa = 9.87SFRR183 pKa = 11.84TTTVRR188 pKa = 11.84SGEE191 pKa = 4.01NRR193 pKa = 11.84TVNLDD198 pKa = 3.24LDD200 pKa = 3.92KK201 pKa = 11.11TLNQYY206 pKa = 11.22LEE208 pKa = 4.31FLNARR213 pKa = 11.84YY214 pKa = 9.54GLSEE218 pKa = 4.9PDD220 pKa = 4.12DD221 pKa = 4.65PSDD224 pKa = 3.61GSIEE228 pKa = 3.8ITANLVNPEE237 pKa = 4.18DD238 pKa = 4.04EE239 pKa = 4.93EE240 pKa = 5.35IEE242 pKa = 4.17LTADD246 pKa = 3.31AEE248 pKa = 4.64VNLDD252 pKa = 4.27EE253 pKa = 5.67ISEE256 pKa = 4.35LTFTAPEE263 pKa = 5.09GYY265 pKa = 10.1DD266 pKa = 3.39SYY268 pKa = 11.78EE269 pKa = 3.66WTVNNMEE276 pKa = 4.34GWDD279 pKa = 3.78NSNTITISEE288 pKa = 4.36SEE290 pKa = 3.93ISDD293 pKa = 3.85VIFPGLNTVDD303 pKa = 4.97LEE305 pKa = 4.47VEE307 pKa = 4.12LGGEE311 pKa = 4.61VYY313 pKa = 10.64HH314 pKa = 6.34GTFTFTAIEE323 pKa = 4.46TILGEE328 pKa = 4.15LVIEE332 pKa = 5.19AILVNPEE339 pKa = 4.27DD340 pKa = 3.85EE341 pKa = 5.07QITLDD346 pKa = 4.15ADD348 pKa = 3.7AEE350 pKa = 4.57VNLDD354 pKa = 4.27EE355 pKa = 5.67ISEE358 pKa = 4.35LTFTAPEE365 pKa = 5.21GYY367 pKa = 10.18DD368 pKa = 3.41SYY370 pKa = 12.02NWTVNNMEE378 pKa = 4.57DD379 pKa = 3.16WGNSNTITISEE390 pKa = 4.17SDD392 pKa = 3.2ISDD395 pKa = 3.73VIFPGLNTVDD405 pKa = 4.97LEE407 pKa = 4.47VEE409 pKa = 4.12LGGEE413 pKa = 4.61VYY415 pKa = 10.64HH416 pKa = 6.34GTFTFTAIEE425 pKa = 4.46TILGEE430 pKa = 4.15LVIEE434 pKa = 5.19AILVNPEE441 pKa = 3.96DD442 pKa = 4.07EE443 pKa = 4.9EE444 pKa = 5.27IEE446 pKa = 4.16LTADD450 pKa = 3.3TEE452 pKa = 4.58VNIDD456 pKa = 4.43EE457 pKa = 4.32ITEE460 pKa = 4.19LTFTAPTGYY469 pKa = 10.44DD470 pKa = 3.17SYY472 pKa = 11.29EE473 pKa = 3.71WTVNDD478 pKa = 3.72MEE480 pKa = 4.9GWDD483 pKa = 3.83NSNTITIPEE492 pKa = 4.1SDD494 pKa = 3.27ISDD497 pKa = 3.99VIKK500 pKa = 10.84LGQNTVDD507 pKa = 4.13LVVEE511 pKa = 4.27KK512 pKa = 10.98AGALYY517 pKa = 9.69SHH519 pKa = 7.48RR520 pKa = 11.84FTFTAISDD528 pKa = 3.62
Molecular weight: 58.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1N6RXP6|A0A1N6RXP6_9SPIO Alpha amylase catalytic domain OS=Alkalispirochaeta americana OX=159291 GN=SAMN05920897_10755 PE=4 SV=1
MM1 pKa = 7.9PPQNIYY7 pKa = 10.07PCYY10 pKa = 9.66GGQGNILVGTAQTAGYY26 pKa = 8.74FVHH29 pKa = 6.51PHH31 pKa = 6.51PFGSQEE37 pKa = 3.72KK38 pKa = 10.05LKK40 pKa = 10.78IEE42 pKa = 4.86GYY44 pKa = 8.84PPKK47 pKa = 10.86AEE49 pKa = 4.33GPGDD53 pKa = 3.92ALRR56 pKa = 11.84DD57 pKa = 3.9VPDD60 pKa = 4.31LPGPAPGKK68 pKa = 9.68ARR70 pKa = 11.84GVAVCQAHH78 pKa = 6.07AQALTARR85 pKa = 11.84EE86 pKa = 4.23VGRR89 pKa = 11.84EE90 pKa = 4.31GYY92 pKa = 8.86PVRR95 pKa = 11.84HH96 pKa = 6.07GPDD99 pKa = 3.45LGLCPLPGRR108 pKa = 11.84PPAEE112 pKa = 3.73GHH114 pKa = 6.59LPGGVVFFHH123 pKa = 6.45QEE125 pKa = 3.0ARR127 pKa = 11.84LPGKK131 pKa = 10.47GEE133 pKa = 4.12SPGPMVQKK141 pKa = 10.83LPTIVDD147 pKa = 3.36NRR149 pKa = 11.84NAPLALAVQGFYY161 pKa = 11.33YY162 pKa = 9.89PGKK165 pKa = 9.83AQLRR169 pKa = 11.84QPLLIPPRR177 pKa = 11.84GVITQNRR184 pKa = 11.84PGNGGNTGMAKK195 pKa = 10.31PLFHH199 pKa = 7.1VSLVGNKK206 pKa = 8.77QSLPGRR212 pKa = 11.84KK213 pKa = 8.97RR214 pKa = 11.84IGEE217 pKa = 4.07PPILRR222 pKa = 11.84SPGCGIHH229 pKa = 6.53GLVGPGRR236 pKa = 11.84HH237 pKa = 5.39NCIDD241 pKa = 3.46SQLPCRR247 pKa = 11.84PGDD250 pKa = 3.08RR251 pKa = 11.84SMIEE255 pKa = 3.67GRR257 pKa = 11.84GLHH260 pKa = 6.02IAISQDD266 pKa = 2.88PPRR269 pKa = 11.84RR270 pKa = 11.84VFSLIGQDD278 pKa = 3.4HH279 pKa = 6.29QKK281 pKa = 10.24PLPAGPLQNPQLAATTPQNHH301 pKa = 6.33KK302 pKa = 9.71PGPGEE307 pKa = 3.91RR308 pKa = 11.84APGTARR314 pKa = 11.84NPPQGPQRR322 pKa = 11.84PPIEE326 pKa = 4.11QEE328 pKa = 3.78QPVYY332 pKa = 10.45LQEE335 pKa = 4.17PRR337 pKa = 11.84QGIAAATRR345 pKa = 11.84GSAPGRR351 pKa = 11.84GSAAAALVVATRR363 pKa = 11.84GSAAAFLVVATRR375 pKa = 11.84GSAAALVVATRR386 pKa = 11.84GSAAGLVVATRR397 pKa = 11.84GSAAALVVATRR408 pKa = 11.84GSAAAFLVVATRR420 pKa = 11.84GSMAASLVVATRR432 pKa = 11.84GSAAALVVATRR443 pKa = 11.84GSTPGRR449 pKa = 11.84GSAASLVVATRR460 pKa = 11.84GSAAAFLVVATRR472 pKa = 11.84GSTTGRR478 pKa = 11.84GSAAAFLVVATRR490 pKa = 11.84GSAAALVVATRR501 pKa = 11.84GSAAALVVATRR512 pKa = 11.84GSMAAFLVVATRR524 pKa = 11.84GSAAALVVATRR535 pKa = 11.84GSAAALVVATRR546 pKa = 11.84GSAAALVVATRR557 pKa = 11.84GSAAGRR563 pKa = 11.84GSAAASLVVATRR575 pKa = 11.84GSAAALVVATRR586 pKa = 11.84GSTPGRR592 pKa = 11.84GSAAAFLVVATRR604 pKa = 11.84GSAAGRR610 pKa = 11.84GSAPGRR616 pKa = 11.84GSAAALVVATRR627 pKa = 11.84GSTTGRR633 pKa = 11.84GSAAAFLVVATRR645 pKa = 11.84GSAAALVVATRR656 pKa = 11.84GSAAAFLATRR666 pKa = 11.84PLAVAHH672 pKa = 6.51RR673 pKa = 11.84RR674 pKa = 11.84RR675 pKa = 11.84NRR677 pKa = 11.84PEE679 pKa = 3.82LQEE682 pKa = 3.79HH683 pKa = 5.68RR684 pKa = 11.84RR685 pKa = 11.84KK686 pKa = 9.31IRR688 pKa = 11.84KK689 pKa = 8.48RR690 pKa = 11.84HH691 pKa = 5.4GPLQRR696 pKa = 11.84PHH698 pKa = 5.72QQLPPKK704 pKa = 9.41HH705 pKa = 6.55AKK707 pKa = 7.76TAHH710 pKa = 6.07GNAPGQPQKK719 pKa = 11.3KK720 pKa = 8.24MKK722 pKa = 9.5QKK724 pKa = 10.05RR725 pKa = 11.84VQPRR729 pKa = 11.84SQPPHH734 pKa = 6.62PPGLSARR741 pKa = 11.84NTVDD745 pKa = 2.88QRR747 pKa = 11.84TARR750 pKa = 11.84HH751 pKa = 6.04LLQTAAQEE759 pKa = 4.22RR760 pKa = 11.84NHH762 pKa = 6.75LGRR765 pKa = 11.84VGPIIVQRR773 pKa = 11.84NHH775 pKa = 5.44QLPPRR780 pKa = 11.84RR781 pKa = 11.84GHH783 pKa = 6.6PRR785 pKa = 11.84QSSPDD790 pKa = 3.55LPCPSGHH797 pKa = 5.98QNQPNPGPGHH807 pKa = 7.25PGQSLPVNRR816 pKa = 11.84ITPVVNDD823 pKa = 3.39NHH825 pKa = 6.79LAPASLGQNRR835 pKa = 11.84PHH837 pKa = 7.37PGHH840 pKa = 6.99KK841 pKa = 9.69PGQTNTPVQGRR852 pKa = 11.84NNQRR856 pKa = 11.84KK857 pKa = 8.97KK858 pKa = 10.23RR859 pKa = 11.84SSVHH863 pKa = 6.12GNHH866 pKa = 5.83STLMHH871 pKa = 7.5PDD873 pKa = 2.71QSMCYY878 pKa = 8.08TFSMDD883 pKa = 3.14HH884 pKa = 6.07ATDD887 pKa = 3.7LVLRR891 pKa = 11.84NPDD894 pKa = 3.54YY895 pKa = 11.32LLTPQGPITGQDD907 pKa = 2.83LWISRR912 pKa = 11.84GAITAILPQGSPPPPGTPTPRR933 pKa = 11.84SVDD936 pKa = 3.49CRR938 pKa = 11.84GKK940 pKa = 10.5AAIPGFKK947 pKa = 9.79NAHH950 pKa = 4.42THH952 pKa = 6.26AAMTLLRR959 pKa = 11.84GYY961 pKa = 10.48GDD963 pKa = 5.38DD964 pKa = 4.26MPLQAWLQEE973 pKa = 4.42RR974 pKa = 11.84IWPAEE979 pKa = 3.49AHH981 pKa = 5.45MTPEE985 pKa = 4.92DD986 pKa = 4.18IYY988 pKa = 10.77WGTRR992 pKa = 11.84LAALEE997 pKa = 4.27MIKK1000 pKa = 10.59SGTTFANDD1008 pKa = 3.56MYY1010 pKa = 10.59FHH1012 pKa = 7.56PAQAAQAFTDD1022 pKa = 3.36AGIRR1026 pKa = 11.84AALGLALFDD1035 pKa = 4.59FDD1037 pKa = 5.04DD1038 pKa = 3.74PHH1040 pKa = 8.4RR1041 pKa = 11.84RR1042 pKa = 11.84AQVQDD1047 pKa = 3.42RR1048 pKa = 11.84LQDD1051 pKa = 3.53NLEE1054 pKa = 4.47TIWPNRR1060 pKa = 11.84RR1061 pKa = 11.84KK1062 pKa = 10.22SPPEE1066 pKa = 3.51EE1067 pKa = 4.33PEE1069 pKa = 3.79QVFPVIAPHH1078 pKa = 7.35SIYY1081 pKa = 10.08TCSGEE1086 pKa = 3.97LLQWCARR1093 pKa = 11.84QASEE1097 pKa = 4.16NHH1099 pKa = 6.09LLYY1102 pKa = 10.1HH1103 pKa = 6.09IHH1105 pKa = 5.84MNEE1108 pKa = 3.45TRR1110 pKa = 11.84RR1111 pKa = 11.84EE1112 pKa = 3.88VDD1114 pKa = 2.54EE1115 pKa = 4.53CLAARR1120 pKa = 11.84GVPPMEE1126 pKa = 4.56HH1127 pKa = 6.04LQNLGVLEE1135 pKa = 4.28TMGDD1139 pKa = 3.46RR1140 pKa = 11.84VVAAHH1145 pKa = 6.17MVWLDD1150 pKa = 3.5PPEE1153 pKa = 5.71LDD1155 pKa = 3.67IAARR1159 pKa = 11.84WGITAVHH1166 pKa = 6.19NPASNMKK1173 pKa = 9.58LASGCFPWQDD1183 pKa = 3.34YY1184 pKa = 10.76ARR1186 pKa = 11.84RR1187 pKa = 11.84KK1188 pKa = 9.5APLMLATDD1196 pKa = 4.65GTASNNNLDD1205 pKa = 4.2MFDD1208 pKa = 3.8EE1209 pKa = 4.56MKK1211 pKa = 10.73LAALLQKK1218 pKa = 10.43HH1219 pKa = 6.18HH1220 pKa = 6.53FQDD1223 pKa = 3.87STRR1226 pKa = 11.84LPAEE1230 pKa = 4.6EE1231 pKa = 4.4ILAIATGARR1240 pKa = 11.84STVFEE1245 pKa = 4.04PWGVGGALTEE1255 pKa = 4.29GAPADD1260 pKa = 3.54IALIDD1265 pKa = 4.3LDD1267 pKa = 4.39HH1268 pKa = 6.68PQMVPVHH1275 pKa = 6.04NLQSNLVYY1283 pKa = 10.35SANGSVVDD1291 pKa = 4.65SVICNGRR1298 pKa = 11.84VLMEE1302 pKa = 3.96NRR1304 pKa = 11.84QVPGEE1309 pKa = 4.14EE1310 pKa = 3.73EE1311 pKa = 4.11VLRR1314 pKa = 11.84EE1315 pKa = 3.89ARR1317 pKa = 11.84RR1318 pKa = 11.84CTRR1321 pKa = 11.84EE1322 pKa = 3.75LVKK1325 pKa = 10.57RR1326 pKa = 11.84AAPP1329 pKa = 3.58
MM1 pKa = 7.9PPQNIYY7 pKa = 10.07PCYY10 pKa = 9.66GGQGNILVGTAQTAGYY26 pKa = 8.74FVHH29 pKa = 6.51PHH31 pKa = 6.51PFGSQEE37 pKa = 3.72KK38 pKa = 10.05LKK40 pKa = 10.78IEE42 pKa = 4.86GYY44 pKa = 8.84PPKK47 pKa = 10.86AEE49 pKa = 4.33GPGDD53 pKa = 3.92ALRR56 pKa = 11.84DD57 pKa = 3.9VPDD60 pKa = 4.31LPGPAPGKK68 pKa = 9.68ARR70 pKa = 11.84GVAVCQAHH78 pKa = 6.07AQALTARR85 pKa = 11.84EE86 pKa = 4.23VGRR89 pKa = 11.84EE90 pKa = 4.31GYY92 pKa = 8.86PVRR95 pKa = 11.84HH96 pKa = 6.07GPDD99 pKa = 3.45LGLCPLPGRR108 pKa = 11.84PPAEE112 pKa = 3.73GHH114 pKa = 6.59LPGGVVFFHH123 pKa = 6.45QEE125 pKa = 3.0ARR127 pKa = 11.84LPGKK131 pKa = 10.47GEE133 pKa = 4.12SPGPMVQKK141 pKa = 10.83LPTIVDD147 pKa = 3.36NRR149 pKa = 11.84NAPLALAVQGFYY161 pKa = 11.33YY162 pKa = 9.89PGKK165 pKa = 9.83AQLRR169 pKa = 11.84QPLLIPPRR177 pKa = 11.84GVITQNRR184 pKa = 11.84PGNGGNTGMAKK195 pKa = 10.31PLFHH199 pKa = 7.1VSLVGNKK206 pKa = 8.77QSLPGRR212 pKa = 11.84KK213 pKa = 8.97RR214 pKa = 11.84IGEE217 pKa = 4.07PPILRR222 pKa = 11.84SPGCGIHH229 pKa = 6.53GLVGPGRR236 pKa = 11.84HH237 pKa = 5.39NCIDD241 pKa = 3.46SQLPCRR247 pKa = 11.84PGDD250 pKa = 3.08RR251 pKa = 11.84SMIEE255 pKa = 3.67GRR257 pKa = 11.84GLHH260 pKa = 6.02IAISQDD266 pKa = 2.88PPRR269 pKa = 11.84RR270 pKa = 11.84VFSLIGQDD278 pKa = 3.4HH279 pKa = 6.29QKK281 pKa = 10.24PLPAGPLQNPQLAATTPQNHH301 pKa = 6.33KK302 pKa = 9.71PGPGEE307 pKa = 3.91RR308 pKa = 11.84APGTARR314 pKa = 11.84NPPQGPQRR322 pKa = 11.84PPIEE326 pKa = 4.11QEE328 pKa = 3.78QPVYY332 pKa = 10.45LQEE335 pKa = 4.17PRR337 pKa = 11.84QGIAAATRR345 pKa = 11.84GSAPGRR351 pKa = 11.84GSAAAALVVATRR363 pKa = 11.84GSAAAFLVVATRR375 pKa = 11.84GSAAALVVATRR386 pKa = 11.84GSAAGLVVATRR397 pKa = 11.84GSAAALVVATRR408 pKa = 11.84GSAAAFLVVATRR420 pKa = 11.84GSMAASLVVATRR432 pKa = 11.84GSAAALVVATRR443 pKa = 11.84GSTPGRR449 pKa = 11.84GSAASLVVATRR460 pKa = 11.84GSAAAFLVVATRR472 pKa = 11.84GSTTGRR478 pKa = 11.84GSAAAFLVVATRR490 pKa = 11.84GSAAALVVATRR501 pKa = 11.84GSAAALVVATRR512 pKa = 11.84GSMAAFLVVATRR524 pKa = 11.84GSAAALVVATRR535 pKa = 11.84GSAAALVVATRR546 pKa = 11.84GSAAALVVATRR557 pKa = 11.84GSAAGRR563 pKa = 11.84GSAAASLVVATRR575 pKa = 11.84GSAAALVVATRR586 pKa = 11.84GSTPGRR592 pKa = 11.84GSAAAFLVVATRR604 pKa = 11.84GSAAGRR610 pKa = 11.84GSAPGRR616 pKa = 11.84GSAAALVVATRR627 pKa = 11.84GSTTGRR633 pKa = 11.84GSAAAFLVVATRR645 pKa = 11.84GSAAALVVATRR656 pKa = 11.84GSAAAFLATRR666 pKa = 11.84PLAVAHH672 pKa = 6.51RR673 pKa = 11.84RR674 pKa = 11.84RR675 pKa = 11.84NRR677 pKa = 11.84PEE679 pKa = 3.82LQEE682 pKa = 3.79HH683 pKa = 5.68RR684 pKa = 11.84RR685 pKa = 11.84KK686 pKa = 9.31IRR688 pKa = 11.84KK689 pKa = 8.48RR690 pKa = 11.84HH691 pKa = 5.4GPLQRR696 pKa = 11.84PHH698 pKa = 5.72QQLPPKK704 pKa = 9.41HH705 pKa = 6.55AKK707 pKa = 7.76TAHH710 pKa = 6.07GNAPGQPQKK719 pKa = 11.3KK720 pKa = 8.24MKK722 pKa = 9.5QKK724 pKa = 10.05RR725 pKa = 11.84VQPRR729 pKa = 11.84SQPPHH734 pKa = 6.62PPGLSARR741 pKa = 11.84NTVDD745 pKa = 2.88QRR747 pKa = 11.84TARR750 pKa = 11.84HH751 pKa = 6.04LLQTAAQEE759 pKa = 4.22RR760 pKa = 11.84NHH762 pKa = 6.75LGRR765 pKa = 11.84VGPIIVQRR773 pKa = 11.84NHH775 pKa = 5.44QLPPRR780 pKa = 11.84RR781 pKa = 11.84GHH783 pKa = 6.6PRR785 pKa = 11.84QSSPDD790 pKa = 3.55LPCPSGHH797 pKa = 5.98QNQPNPGPGHH807 pKa = 7.25PGQSLPVNRR816 pKa = 11.84ITPVVNDD823 pKa = 3.39NHH825 pKa = 6.79LAPASLGQNRR835 pKa = 11.84PHH837 pKa = 7.37PGHH840 pKa = 6.99KK841 pKa = 9.69PGQTNTPVQGRR852 pKa = 11.84NNQRR856 pKa = 11.84KK857 pKa = 8.97KK858 pKa = 10.23RR859 pKa = 11.84SSVHH863 pKa = 6.12GNHH866 pKa = 5.83STLMHH871 pKa = 7.5PDD873 pKa = 2.71QSMCYY878 pKa = 8.08TFSMDD883 pKa = 3.14HH884 pKa = 6.07ATDD887 pKa = 3.7LVLRR891 pKa = 11.84NPDD894 pKa = 3.54YY895 pKa = 11.32LLTPQGPITGQDD907 pKa = 2.83LWISRR912 pKa = 11.84GAITAILPQGSPPPPGTPTPRR933 pKa = 11.84SVDD936 pKa = 3.49CRR938 pKa = 11.84GKK940 pKa = 10.5AAIPGFKK947 pKa = 9.79NAHH950 pKa = 4.42THH952 pKa = 6.26AAMTLLRR959 pKa = 11.84GYY961 pKa = 10.48GDD963 pKa = 5.38DD964 pKa = 4.26MPLQAWLQEE973 pKa = 4.42RR974 pKa = 11.84IWPAEE979 pKa = 3.49AHH981 pKa = 5.45MTPEE985 pKa = 4.92DD986 pKa = 4.18IYY988 pKa = 10.77WGTRR992 pKa = 11.84LAALEE997 pKa = 4.27MIKK1000 pKa = 10.59SGTTFANDD1008 pKa = 3.56MYY1010 pKa = 10.59FHH1012 pKa = 7.56PAQAAQAFTDD1022 pKa = 3.36AGIRR1026 pKa = 11.84AALGLALFDD1035 pKa = 4.59FDD1037 pKa = 5.04DD1038 pKa = 3.74PHH1040 pKa = 8.4RR1041 pKa = 11.84RR1042 pKa = 11.84AQVQDD1047 pKa = 3.42RR1048 pKa = 11.84LQDD1051 pKa = 3.53NLEE1054 pKa = 4.47TIWPNRR1060 pKa = 11.84RR1061 pKa = 11.84KK1062 pKa = 10.22SPPEE1066 pKa = 3.51EE1067 pKa = 4.33PEE1069 pKa = 3.79QVFPVIAPHH1078 pKa = 7.35SIYY1081 pKa = 10.08TCSGEE1086 pKa = 3.97LLQWCARR1093 pKa = 11.84QASEE1097 pKa = 4.16NHH1099 pKa = 6.09LLYY1102 pKa = 10.1HH1103 pKa = 6.09IHH1105 pKa = 5.84MNEE1108 pKa = 3.45TRR1110 pKa = 11.84RR1111 pKa = 11.84EE1112 pKa = 3.88VDD1114 pKa = 2.54EE1115 pKa = 4.53CLAARR1120 pKa = 11.84GVPPMEE1126 pKa = 4.56HH1127 pKa = 6.04LQNLGVLEE1135 pKa = 4.28TMGDD1139 pKa = 3.46RR1140 pKa = 11.84VVAAHH1145 pKa = 6.17MVWLDD1150 pKa = 3.5PPEE1153 pKa = 5.71LDD1155 pKa = 3.67IAARR1159 pKa = 11.84WGITAVHH1166 pKa = 6.19NPASNMKK1173 pKa = 9.58LASGCFPWQDD1183 pKa = 3.34YY1184 pKa = 10.76ARR1186 pKa = 11.84RR1187 pKa = 11.84KK1188 pKa = 9.5APLMLATDD1196 pKa = 4.65GTASNNNLDD1205 pKa = 4.2MFDD1208 pKa = 3.8EE1209 pKa = 4.56MKK1211 pKa = 10.73LAALLQKK1218 pKa = 10.43HH1219 pKa = 6.18HH1220 pKa = 6.53FQDD1223 pKa = 3.87STRR1226 pKa = 11.84LPAEE1230 pKa = 4.6EE1231 pKa = 4.4ILAIATGARR1240 pKa = 11.84STVFEE1245 pKa = 4.04PWGVGGALTEE1255 pKa = 4.29GAPADD1260 pKa = 3.54IALIDD1265 pKa = 4.3LDD1267 pKa = 4.39HH1268 pKa = 6.68PQMVPVHH1275 pKa = 6.04NLQSNLVYY1283 pKa = 10.35SANGSVVDD1291 pKa = 4.65SVICNGRR1298 pKa = 11.84VLMEE1302 pKa = 3.96NRR1304 pKa = 11.84QVPGEE1309 pKa = 4.14EE1310 pKa = 3.73EE1311 pKa = 4.11VLRR1314 pKa = 11.84EE1315 pKa = 3.89ARR1317 pKa = 11.84RR1318 pKa = 11.84CTRR1321 pKa = 11.84EE1322 pKa = 3.75LVKK1325 pKa = 10.57RR1326 pKa = 11.84AAPP1329 pKa = 3.58
Molecular weight: 139.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1021938 |
26 |
2692 |
353.5 |
39.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.107 ± 0.047 | 0.84 ± 0.015 |
5.402 ± 0.038 | 7.207 ± 0.047 |
4.044 ± 0.033 | 8.085 ± 0.046 |
2.071 ± 0.021 | 5.593 ± 0.035 |
2.981 ± 0.038 | 10.911 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.142 ± 0.024 | 2.716 ± 0.028 |
5.058 ± 0.042 | 3.756 ± 0.028 |
7.672 ± 0.047 | 6.287 ± 0.038 |
5.182 ± 0.048 | 7.146 ± 0.045 |
1.155 ± 0.017 | 2.644 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
