
Hubei tombus-like virus 23
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.7
Get precalculated fractions of proteins
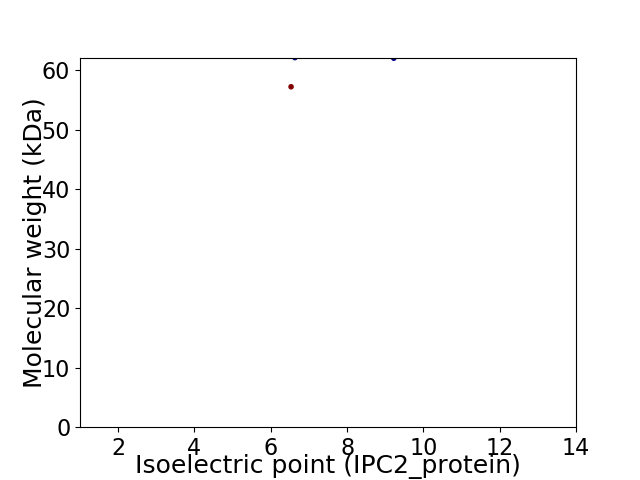
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
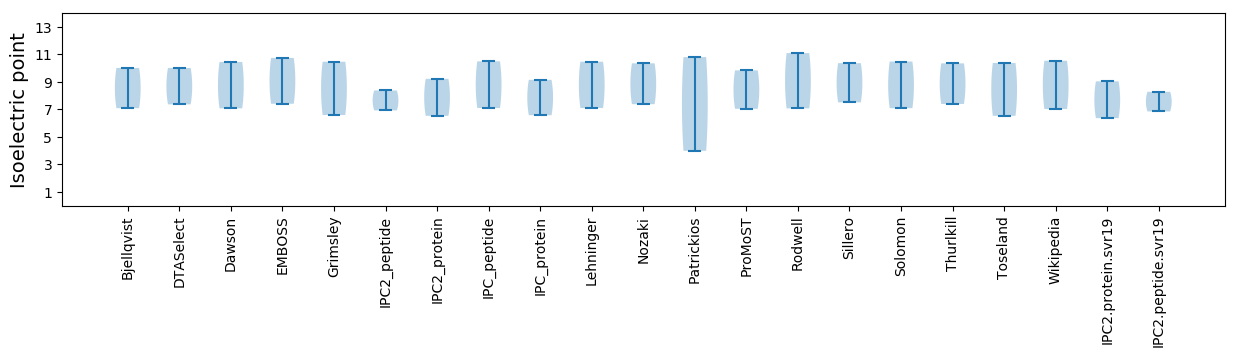
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGK3|A0A1L3KGK3_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 23 OX=1923270 PE=4 SV=1
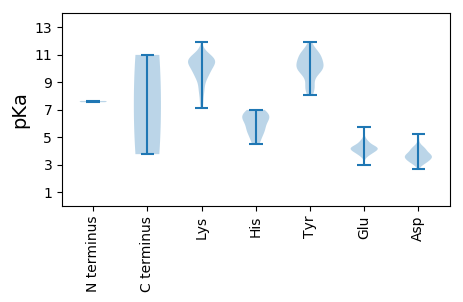
MM1 pKa = 7.62GICLGSLEE9 pKa = 4.17PSKK12 pKa = 10.79LIKK15 pKa = 10.63AKK17 pKa = 10.16ILNIKK22 pKa = 8.72TDD24 pKa = 3.58PCQHH28 pKa = 5.88KK29 pKa = 9.24TYY31 pKa = 9.78IYY33 pKa = 10.06PLINPPPGIDD43 pKa = 3.11HH44 pKa = 5.59QTQVVYY50 pKa = 9.67QPCQLNEE57 pKa = 3.84LHH59 pKa = 6.65SLNKK63 pKa = 9.65RR64 pKa = 11.84HH65 pKa = 6.4LVFHH69 pKa = 6.97KK70 pKa = 9.87WPKK73 pKa = 8.21EE74 pKa = 3.74QLVNEE79 pKa = 5.63AIQHH83 pKa = 5.33TKK85 pKa = 10.37QLLKK89 pKa = 10.85DD90 pKa = 3.86GEE92 pKa = 4.56NVQPITPMEE101 pKa = 3.88YY102 pKa = 10.74AMTRR106 pKa = 11.84KK107 pKa = 8.81GAKK110 pKa = 8.78RR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84YY114 pKa = 8.3QQAAEE119 pKa = 3.92EE120 pKa = 3.85LDD122 pKa = 3.96YY123 pKa = 11.92YY124 pKa = 10.99GMKK127 pKa = 10.36PSATNVTAFIKK138 pKa = 10.05IEE140 pKa = 3.66KK141 pKa = 8.4WDD143 pKa = 3.89RR144 pKa = 11.84EE145 pKa = 4.14VAMVKK150 pKa = 9.67SPRR153 pKa = 11.84MIQYY157 pKa = 8.78RR158 pKa = 11.84HH159 pKa = 5.45PTYY162 pKa = 10.87VSTIASRR169 pKa = 11.84LYY171 pKa = 10.14PFEE174 pKa = 4.27EE175 pKa = 4.41KK176 pKa = 10.41LWEE179 pKa = 4.15TEE181 pKa = 4.03RR182 pKa = 11.84NGCRR186 pKa = 11.84VFAKK190 pKa = 10.65GLDD193 pKa = 3.8SYY195 pKa = 10.07QTAAQLRR202 pKa = 11.84MKK204 pKa = 9.96WEE206 pKa = 4.01RR207 pKa = 11.84CDD209 pKa = 3.57RR210 pKa = 11.84PIAIMKK216 pKa = 9.83DD217 pKa = 3.45FSKK220 pKa = 10.71FDD222 pKa = 3.56SCITLPWIKK231 pKa = 10.47GEE233 pKa = 4.05KK234 pKa = 9.54EE235 pKa = 3.62IYY237 pKa = 10.05EE238 pKa = 4.17LTGTMDD244 pKa = 3.09EE245 pKa = 4.78MDD247 pKa = 3.57YY248 pKa = 11.21QFNNTCSTVNGVSYY262 pKa = 10.49KK263 pKa = 10.71CEE265 pKa = 3.66ARR267 pKa = 11.84KK268 pKa = 9.14MSGEE272 pKa = 4.23YY273 pKa = 8.66NTSCGDD279 pKa = 4.38SIVNWSATDD288 pKa = 3.48KK289 pKa = 10.34AFHH292 pKa = 6.95MITGHH297 pKa = 4.89YY298 pKa = 9.57RR299 pKa = 11.84YY300 pKa = 9.99YY301 pKa = 10.55PNINGDD307 pKa = 4.19DD308 pKa = 3.61DD309 pKa = 5.24VMIVDD314 pKa = 4.12EE315 pKa = 5.06SEE317 pKa = 3.7IGDD320 pKa = 3.59IEE322 pKa = 4.32EE323 pKa = 4.56FLPKK327 pKa = 10.37LIEE330 pKa = 4.14NLEE333 pKa = 4.06LLGFKK338 pKa = 9.8TDD340 pKa = 3.09ASYY343 pKa = 10.61TRR345 pKa = 11.84TFEE348 pKa = 4.68EE349 pKa = 4.91IEE351 pKa = 4.33FCQSQPVEE359 pKa = 4.13VEE361 pKa = 4.11SNIWRR366 pKa = 11.84MIRR369 pKa = 11.84KK370 pKa = 7.14PARR373 pKa = 11.84AISRR377 pKa = 11.84DD378 pKa = 3.65CYY380 pKa = 10.43SVRR383 pKa = 11.84KK384 pKa = 9.95YY385 pKa = 11.06GGTAWYY391 pKa = 10.04RR392 pKa = 11.84LAASLGYY399 pKa = 10.03CEE401 pKa = 4.8YY402 pKa = 11.03ALNDD406 pKa = 4.17GVPVLQSWAKK416 pKa = 9.01YY417 pKa = 8.06LQRR420 pKa = 11.84SSKK423 pKa = 9.73GKK425 pKa = 9.98KK426 pKa = 9.3VLNQEE431 pKa = 2.93IEE433 pKa = 4.11YY434 pKa = 9.97RR435 pKa = 11.84VKK437 pKa = 11.02VEE439 pKa = 4.19IKK441 pKa = 10.67NKK443 pKa = 8.05TKK445 pKa = 10.51YY446 pKa = 8.62RR447 pKa = 11.84TTIDD451 pKa = 3.27PVARR455 pKa = 11.84EE456 pKa = 3.96SFAIAFGIEE465 pKa = 3.91PTQQIAIEE473 pKa = 4.18SWLDD477 pKa = 3.52NQEE480 pKa = 4.24SPEE483 pKa = 4.07LLPYY487 pKa = 10.28SEE489 pKa = 5.71GYY491 pKa = 8.63WGG493 pKa = 3.75
MM1 pKa = 7.62GICLGSLEE9 pKa = 4.17PSKK12 pKa = 10.79LIKK15 pKa = 10.63AKK17 pKa = 10.16ILNIKK22 pKa = 8.72TDD24 pKa = 3.58PCQHH28 pKa = 5.88KK29 pKa = 9.24TYY31 pKa = 9.78IYY33 pKa = 10.06PLINPPPGIDD43 pKa = 3.11HH44 pKa = 5.59QTQVVYY50 pKa = 9.67QPCQLNEE57 pKa = 3.84LHH59 pKa = 6.65SLNKK63 pKa = 9.65RR64 pKa = 11.84HH65 pKa = 6.4LVFHH69 pKa = 6.97KK70 pKa = 9.87WPKK73 pKa = 8.21EE74 pKa = 3.74QLVNEE79 pKa = 5.63AIQHH83 pKa = 5.33TKK85 pKa = 10.37QLLKK89 pKa = 10.85DD90 pKa = 3.86GEE92 pKa = 4.56NVQPITPMEE101 pKa = 3.88YY102 pKa = 10.74AMTRR106 pKa = 11.84KK107 pKa = 8.81GAKK110 pKa = 8.78RR111 pKa = 11.84RR112 pKa = 11.84RR113 pKa = 11.84YY114 pKa = 8.3QQAAEE119 pKa = 3.92EE120 pKa = 3.85LDD122 pKa = 3.96YY123 pKa = 11.92YY124 pKa = 10.99GMKK127 pKa = 10.36PSATNVTAFIKK138 pKa = 10.05IEE140 pKa = 3.66KK141 pKa = 8.4WDD143 pKa = 3.89RR144 pKa = 11.84EE145 pKa = 4.14VAMVKK150 pKa = 9.67SPRR153 pKa = 11.84MIQYY157 pKa = 8.78RR158 pKa = 11.84HH159 pKa = 5.45PTYY162 pKa = 10.87VSTIASRR169 pKa = 11.84LYY171 pKa = 10.14PFEE174 pKa = 4.27EE175 pKa = 4.41KK176 pKa = 10.41LWEE179 pKa = 4.15TEE181 pKa = 4.03RR182 pKa = 11.84NGCRR186 pKa = 11.84VFAKK190 pKa = 10.65GLDD193 pKa = 3.8SYY195 pKa = 10.07QTAAQLRR202 pKa = 11.84MKK204 pKa = 9.96WEE206 pKa = 4.01RR207 pKa = 11.84CDD209 pKa = 3.57RR210 pKa = 11.84PIAIMKK216 pKa = 9.83DD217 pKa = 3.45FSKK220 pKa = 10.71FDD222 pKa = 3.56SCITLPWIKK231 pKa = 10.47GEE233 pKa = 4.05KK234 pKa = 9.54EE235 pKa = 3.62IYY237 pKa = 10.05EE238 pKa = 4.17LTGTMDD244 pKa = 3.09EE245 pKa = 4.78MDD247 pKa = 3.57YY248 pKa = 11.21QFNNTCSTVNGVSYY262 pKa = 10.49KK263 pKa = 10.71CEE265 pKa = 3.66ARR267 pKa = 11.84KK268 pKa = 9.14MSGEE272 pKa = 4.23YY273 pKa = 8.66NTSCGDD279 pKa = 4.38SIVNWSATDD288 pKa = 3.48KK289 pKa = 10.34AFHH292 pKa = 6.95MITGHH297 pKa = 4.89YY298 pKa = 9.57RR299 pKa = 11.84YY300 pKa = 9.99YY301 pKa = 10.55PNINGDD307 pKa = 4.19DD308 pKa = 3.61DD309 pKa = 5.24VMIVDD314 pKa = 4.12EE315 pKa = 5.06SEE317 pKa = 3.7IGDD320 pKa = 3.59IEE322 pKa = 4.32EE323 pKa = 4.56FLPKK327 pKa = 10.37LIEE330 pKa = 4.14NLEE333 pKa = 4.06LLGFKK338 pKa = 9.8TDD340 pKa = 3.09ASYY343 pKa = 10.61TRR345 pKa = 11.84TFEE348 pKa = 4.68EE349 pKa = 4.91IEE351 pKa = 4.33FCQSQPVEE359 pKa = 4.13VEE361 pKa = 4.11SNIWRR366 pKa = 11.84MIRR369 pKa = 11.84KK370 pKa = 7.14PARR373 pKa = 11.84AISRR377 pKa = 11.84DD378 pKa = 3.65CYY380 pKa = 10.43SVRR383 pKa = 11.84KK384 pKa = 9.95YY385 pKa = 11.06GGTAWYY391 pKa = 10.04RR392 pKa = 11.84LAASLGYY399 pKa = 10.03CEE401 pKa = 4.8YY402 pKa = 11.03ALNDD406 pKa = 4.17GVPVLQSWAKK416 pKa = 9.01YY417 pKa = 8.06LQRR420 pKa = 11.84SSKK423 pKa = 9.73GKK425 pKa = 9.98KK426 pKa = 9.3VLNQEE431 pKa = 2.93IEE433 pKa = 4.11YY434 pKa = 9.97RR435 pKa = 11.84VKK437 pKa = 11.02VEE439 pKa = 4.19IKK441 pKa = 10.67NKK443 pKa = 8.05TKK445 pKa = 10.51YY446 pKa = 8.62RR447 pKa = 11.84TTIDD451 pKa = 3.27PVARR455 pKa = 11.84EE456 pKa = 3.96SFAIAFGIEE465 pKa = 3.91PTQQIAIEE473 pKa = 4.18SWLDD477 pKa = 3.52NQEE480 pKa = 4.24SPEE483 pKa = 4.07LLPYY487 pKa = 10.28SEE489 pKa = 5.71GYY491 pKa = 8.63WGG493 pKa = 3.75
Molecular weight: 57.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGK3|A0A1L3KGK3_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 23 OX=1923270 PE=4 SV=1
MM1 pKa = 7.53NDD3 pKa = 3.16IIKK6 pKa = 10.7SKK8 pKa = 10.61EE9 pKa = 3.96EE10 pKa = 3.79PVQDD14 pKa = 3.69ISGTTPSTADD24 pKa = 3.19EE25 pKa = 4.33SRR27 pKa = 11.84ASFGKK32 pKa = 10.66APGEE36 pKa = 4.24GFQSKK41 pKa = 9.88PIQTNQSMKK50 pKa = 10.6KK51 pKa = 8.25NSKK54 pKa = 9.69VNSSQRR60 pKa = 11.84RR61 pKa = 11.84AAEE64 pKa = 4.02RR65 pKa = 11.84KK66 pKa = 9.44NEE68 pKa = 4.05RR69 pKa = 11.84DD70 pKa = 3.36DD71 pKa = 3.59KK72 pKa = 11.87ANVSNHH78 pKa = 4.5AVKK81 pKa = 10.81NGLSGGMRR89 pKa = 11.84FKK91 pKa = 10.96FNKK94 pKa = 9.44KK95 pKa = 9.94SKK97 pKa = 9.04LTYY100 pKa = 9.98DD101 pKa = 3.24QQKK104 pKa = 11.34ANFEE108 pKa = 4.26AMLNPVQKK116 pKa = 10.92ANNNPQDD123 pKa = 4.07FPNKK127 pKa = 7.46WRR129 pKa = 11.84QVAARR134 pKa = 11.84NKK136 pKa = 10.41RR137 pKa = 11.84KK138 pKa = 9.45DD139 pKa = 3.57AKK141 pKa = 10.73SATQRR146 pKa = 11.84QCAFTGRR153 pKa = 11.84QYY155 pKa = 11.11IATNATQRR163 pKa = 11.84LIEE166 pKa = 4.41KK167 pKa = 9.45RR168 pKa = 11.84NQSIHH173 pKa = 6.05HH174 pKa = 6.33LRR176 pKa = 11.84MEE178 pKa = 4.16NQTIRR183 pKa = 11.84RR184 pKa = 11.84EE185 pKa = 4.11LQSLKK190 pKa = 10.85EE191 pKa = 4.44CINDD195 pKa = 3.36LTKK198 pKa = 10.97LLRR201 pKa = 11.84GGKK204 pKa = 7.55QQNLRR209 pKa = 11.84KK210 pKa = 8.47PTLNSTKK217 pKa = 10.38SGEE220 pKa = 4.26SSSKK224 pKa = 10.22KK225 pKa = 10.16SRR227 pKa = 11.84TSEE230 pKa = 3.85VGTMSEE236 pKa = 4.25MNSEE240 pKa = 4.28RR241 pKa = 11.84FTTRR245 pKa = 11.84QSLDD249 pKa = 3.32PLKK252 pKa = 10.09RR253 pKa = 11.84TQSLQCLSYY262 pKa = 11.06KK263 pKa = 10.53DD264 pKa = 4.8ALLKK268 pKa = 10.48HH269 pKa = 6.48LNVKK273 pKa = 9.56PKK275 pKa = 10.56SITRR279 pKa = 11.84TSSLKK284 pKa = 10.97LNTKK288 pKa = 7.28QEE290 pKa = 4.46RR291 pKa = 11.84KK292 pKa = 10.39HH293 pKa = 5.63MMNSTEE299 pKa = 3.9QSTSAMRR306 pKa = 11.84NISSGSEE313 pKa = 3.59RR314 pKa = 11.84NKK316 pKa = 8.18QTKK319 pKa = 9.38GCQKK323 pKa = 10.5AEE325 pKa = 4.25SRR327 pKa = 11.84GHH329 pKa = 4.75QAQNVSTKK337 pKa = 9.76RR338 pKa = 11.84SRR340 pKa = 11.84MTTRR344 pKa = 11.84HH345 pKa = 5.4STDD348 pKa = 2.66GTQVIKK354 pKa = 10.51MEE356 pKa = 4.71SKK358 pKa = 8.42GTAPEE363 pKa = 3.94THH365 pKa = 6.26LTDD368 pKa = 4.2SGMQTEE374 pKa = 4.44EE375 pKa = 3.93VDD377 pKa = 3.18NEE379 pKa = 4.18TRR381 pKa = 11.84DD382 pKa = 3.86TIVDD386 pKa = 3.7DD387 pKa = 4.29VRR389 pKa = 11.84LINQGLSEE397 pKa = 4.64EE398 pKa = 4.6DD399 pKa = 3.87SLPKK403 pKa = 10.49PMIATKK409 pKa = 10.11IANKK413 pKa = 10.11GKK415 pKa = 10.23EE416 pKa = 3.7SGNRR420 pKa = 11.84FKK422 pKa = 11.22RR423 pKa = 11.84NLLKK427 pKa = 10.4QRR429 pKa = 11.84EE430 pKa = 4.31PVLQLPLGEE439 pKa = 4.17IQEE442 pKa = 4.25EE443 pKa = 4.54LISYY447 pKa = 9.52LRR449 pKa = 11.84IKK451 pKa = 10.78AGFHH455 pKa = 6.94AMDD458 pKa = 3.02TMMIRR463 pKa = 11.84RR464 pKa = 11.84LFQHH468 pKa = 6.66AEE470 pKa = 3.61TWADD474 pKa = 3.77KK475 pKa = 10.63YY476 pKa = 11.09CKK478 pKa = 9.64EE479 pKa = 4.63LSWQEE484 pKa = 3.96KK485 pKa = 9.23EE486 pKa = 4.35RR487 pKa = 11.84LIADD491 pKa = 4.09GVVTAMVPSEE501 pKa = 3.88HH502 pKa = 6.46MMVLRR507 pKa = 11.84QKK509 pKa = 11.04LKK511 pKa = 11.08DD512 pKa = 3.34SDD514 pKa = 4.19VQSKK518 pKa = 7.57LHH520 pKa = 6.45KK521 pKa = 10.52ASDD524 pKa = 3.64MAKK527 pKa = 10.45GDD529 pKa = 3.47LGKK532 pKa = 10.81RR533 pKa = 11.84NFFGKK538 pKa = 10.29EE539 pKa = 3.21ISLPTASKK547 pKa = 10.97
MM1 pKa = 7.53NDD3 pKa = 3.16IIKK6 pKa = 10.7SKK8 pKa = 10.61EE9 pKa = 3.96EE10 pKa = 3.79PVQDD14 pKa = 3.69ISGTTPSTADD24 pKa = 3.19EE25 pKa = 4.33SRR27 pKa = 11.84ASFGKK32 pKa = 10.66APGEE36 pKa = 4.24GFQSKK41 pKa = 9.88PIQTNQSMKK50 pKa = 10.6KK51 pKa = 8.25NSKK54 pKa = 9.69VNSSQRR60 pKa = 11.84RR61 pKa = 11.84AAEE64 pKa = 4.02RR65 pKa = 11.84KK66 pKa = 9.44NEE68 pKa = 4.05RR69 pKa = 11.84DD70 pKa = 3.36DD71 pKa = 3.59KK72 pKa = 11.87ANVSNHH78 pKa = 4.5AVKK81 pKa = 10.81NGLSGGMRR89 pKa = 11.84FKK91 pKa = 10.96FNKK94 pKa = 9.44KK95 pKa = 9.94SKK97 pKa = 9.04LTYY100 pKa = 9.98DD101 pKa = 3.24QQKK104 pKa = 11.34ANFEE108 pKa = 4.26AMLNPVQKK116 pKa = 10.92ANNNPQDD123 pKa = 4.07FPNKK127 pKa = 7.46WRR129 pKa = 11.84QVAARR134 pKa = 11.84NKK136 pKa = 10.41RR137 pKa = 11.84KK138 pKa = 9.45DD139 pKa = 3.57AKK141 pKa = 10.73SATQRR146 pKa = 11.84QCAFTGRR153 pKa = 11.84QYY155 pKa = 11.11IATNATQRR163 pKa = 11.84LIEE166 pKa = 4.41KK167 pKa = 9.45RR168 pKa = 11.84NQSIHH173 pKa = 6.05HH174 pKa = 6.33LRR176 pKa = 11.84MEE178 pKa = 4.16NQTIRR183 pKa = 11.84RR184 pKa = 11.84EE185 pKa = 4.11LQSLKK190 pKa = 10.85EE191 pKa = 4.44CINDD195 pKa = 3.36LTKK198 pKa = 10.97LLRR201 pKa = 11.84GGKK204 pKa = 7.55QQNLRR209 pKa = 11.84KK210 pKa = 8.47PTLNSTKK217 pKa = 10.38SGEE220 pKa = 4.26SSSKK224 pKa = 10.22KK225 pKa = 10.16SRR227 pKa = 11.84TSEE230 pKa = 3.85VGTMSEE236 pKa = 4.25MNSEE240 pKa = 4.28RR241 pKa = 11.84FTTRR245 pKa = 11.84QSLDD249 pKa = 3.32PLKK252 pKa = 10.09RR253 pKa = 11.84TQSLQCLSYY262 pKa = 11.06KK263 pKa = 10.53DD264 pKa = 4.8ALLKK268 pKa = 10.48HH269 pKa = 6.48LNVKK273 pKa = 9.56PKK275 pKa = 10.56SITRR279 pKa = 11.84TSSLKK284 pKa = 10.97LNTKK288 pKa = 7.28QEE290 pKa = 4.46RR291 pKa = 11.84KK292 pKa = 10.39HH293 pKa = 5.63MMNSTEE299 pKa = 3.9QSTSAMRR306 pKa = 11.84NISSGSEE313 pKa = 3.59RR314 pKa = 11.84NKK316 pKa = 8.18QTKK319 pKa = 9.38GCQKK323 pKa = 10.5AEE325 pKa = 4.25SRR327 pKa = 11.84GHH329 pKa = 4.75QAQNVSTKK337 pKa = 9.76RR338 pKa = 11.84SRR340 pKa = 11.84MTTRR344 pKa = 11.84HH345 pKa = 5.4STDD348 pKa = 2.66GTQVIKK354 pKa = 10.51MEE356 pKa = 4.71SKK358 pKa = 8.42GTAPEE363 pKa = 3.94THH365 pKa = 6.26LTDD368 pKa = 4.2SGMQTEE374 pKa = 4.44EE375 pKa = 3.93VDD377 pKa = 3.18NEE379 pKa = 4.18TRR381 pKa = 11.84DD382 pKa = 3.86TIVDD386 pKa = 3.7DD387 pKa = 4.29VRR389 pKa = 11.84LINQGLSEE397 pKa = 4.64EE398 pKa = 4.6DD399 pKa = 3.87SLPKK403 pKa = 10.49PMIATKK409 pKa = 10.11IANKK413 pKa = 10.11GKK415 pKa = 10.23EE416 pKa = 3.7SGNRR420 pKa = 11.84FKK422 pKa = 11.22RR423 pKa = 11.84NLLKK427 pKa = 10.4QRR429 pKa = 11.84EE430 pKa = 4.31PVLQLPLGEE439 pKa = 4.17IQEE442 pKa = 4.25EE443 pKa = 4.54LISYY447 pKa = 9.52LRR449 pKa = 11.84IKK451 pKa = 10.78AGFHH455 pKa = 6.94AMDD458 pKa = 3.02TMMIRR463 pKa = 11.84RR464 pKa = 11.84LFQHH468 pKa = 6.66AEE470 pKa = 3.61TWADD474 pKa = 3.77KK475 pKa = 10.63YY476 pKa = 11.09CKK478 pKa = 9.64EE479 pKa = 4.63LSWQEE484 pKa = 3.96KK485 pKa = 9.23EE486 pKa = 4.35RR487 pKa = 11.84LIADD491 pKa = 4.09GVVTAMVPSEE501 pKa = 3.88HH502 pKa = 6.46MMVLRR507 pKa = 11.84QKK509 pKa = 11.04LKK511 pKa = 11.08DD512 pKa = 3.34SDD514 pKa = 4.19VQSKK518 pKa = 7.57LHH520 pKa = 6.45KK521 pKa = 10.52ASDD524 pKa = 3.64MAKK527 pKa = 10.45GDD529 pKa = 3.47LGKK532 pKa = 10.81RR533 pKa = 11.84NFFGKK538 pKa = 10.29EE539 pKa = 3.21ISLPTASKK547 pKa = 10.97
Molecular weight: 62.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1040 |
493 |
547 |
520.0 |
59.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.962 ± 0.05 | 1.635 ± 0.507 |
4.615 ± 0.032 | 7.596 ± 0.586 |
2.596 ± 0.155 | 5.0 ± 0.045 |
2.019 ± 0.123 | 5.577 ± 1.094 |
9.423 ± 1.088 | 7.019 ± 0.207 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.365 ± 0.333 | 5.385 ± 0.714 |
4.038 ± 0.655 | 5.865 ± 0.761 |
6.538 ± 0.674 | 8.077 ± 1.264 |
6.538 ± 0.545 | 4.135 ± 0.465 |
1.346 ± 0.561 | 3.269 ± 1.658 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
