
Apis mellifera associated microvirus 9
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
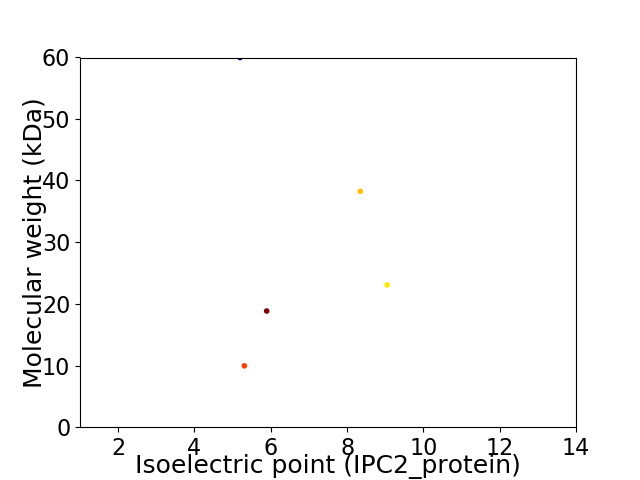
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UTX7|A0A3S8UTX7_9VIRU DNA pilot protein OS=Apis mellifera associated microvirus 9 OX=2494796 PE=4 SV=1
MM1 pKa = 7.27HH2 pKa = 7.49ASNYY6 pKa = 9.99KK7 pKa = 8.94MPSNMTHH14 pKa = 5.99NFSRR18 pKa = 11.84VPQVHH23 pKa = 5.96NPRR26 pKa = 11.84STFVSPYY33 pKa = 8.78TYY35 pKa = 9.28KK36 pKa = 10.25TALNAGFLYY45 pKa = 10.06PFFMDD50 pKa = 3.71EE51 pKa = 4.4VLPGDD56 pKa = 3.99TFNDD60 pKa = 3.61DD61 pKa = 3.38FTVFARR67 pKa = 11.84LTTPIKK73 pKa = 10.43PIMDD77 pKa = 4.32NLYY80 pKa = 10.07LDD82 pKa = 3.56TFFFYY87 pKa = 10.39IPNRR91 pKa = 11.84LLDD94 pKa = 4.11DD95 pKa = 3.29TWEE98 pKa = 4.35KK99 pKa = 11.43LNGAQDD105 pKa = 3.75NPGDD109 pKa = 4.17SIDD112 pKa = 3.84YY113 pKa = 9.17VVPQIPAPAVTGFASEE129 pKa = 4.43SLFDD133 pKa = 3.83YY134 pKa = 10.35MGIRR138 pKa = 11.84PGVTNFSIDD147 pKa = 3.18AYY149 pKa = 10.98HH150 pKa = 6.88PLAYY154 pKa = 10.57NLLFNTNFRR163 pKa = 11.84DD164 pKa = 3.47EE165 pKa = 4.24NLQDD169 pKa = 3.11SVYY172 pKa = 10.06FSKK175 pKa = 11.24GPGPHH180 pKa = 6.98AATNFNLLRR189 pKa = 11.84RR190 pKa = 11.84GKK192 pKa = 10.18RR193 pKa = 11.84FDD195 pKa = 3.87YY196 pKa = 8.15FTSCLPFPQKK206 pKa = 10.69GPEE209 pKa = 3.76VTIPILGDD217 pKa = 3.34APLIGIGTKK226 pKa = 9.98TNVGFSPGVTGYY238 pKa = 10.75DD239 pKa = 3.14YY240 pKa = 11.43DD241 pKa = 3.91GAVTYY246 pKa = 8.07TQGRR250 pKa = 11.84ATDD253 pKa = 3.58AGGANNAIIVDD264 pKa = 3.87VDD266 pKa = 3.71PATGLPKK273 pKa = 9.73WAASMAEE280 pKa = 3.91VSGVSINQFRR290 pKa = 11.84TSYY293 pKa = 9.7QIQMLLEE300 pKa = 4.04NDD302 pKa = 3.13ARR304 pKa = 11.84GGTRR308 pKa = 11.84YY309 pKa = 9.96PEE311 pKa = 3.8ILRR314 pKa = 11.84AHH316 pKa = 6.7FNVISPDD323 pKa = 3.24FRR325 pKa = 11.84LQRR328 pKa = 11.84PEE330 pKa = 3.9YY331 pKa = 10.47LGGTSVPVTVTPVPQTSSTDD351 pKa = 3.27NTSPQGNLAAYY362 pKa = 7.08ATSLNRR368 pKa = 11.84CGYY371 pKa = 7.33TKK373 pKa = 10.66SFVEE377 pKa = 4.15HH378 pKa = 6.35GVILGLCSIRR388 pKa = 11.84ADD390 pKa = 3.38LTYY393 pKa = 10.61QYY395 pKa = 11.07GLRR398 pKa = 11.84RR399 pKa = 11.84MWSRR403 pKa = 11.84QTRR406 pKa = 11.84YY407 pKa = 10.68DD408 pKa = 3.82FYY410 pKa = 10.82WPAFAHH416 pKa = 5.97LGEE419 pKa = 4.15MAVLNKK425 pKa = 10.06EE426 pKa = 4.09IYY428 pKa = 9.82IQGTADD434 pKa = 3.5DD435 pKa = 4.53NLVFGYY441 pKa = 10.16QEE443 pKa = 3.16AWADD447 pKa = 3.88YY448 pKa = 9.56KK449 pKa = 10.59YY450 pKa = 10.56HH451 pKa = 7.31PSLITGKK458 pKa = 8.75MRR460 pKa = 11.84SQDD463 pKa = 3.81PQSLDD468 pKa = 2.74IWHH471 pKa = 7.5LSQEE475 pKa = 4.43FQSLPLLNDD484 pKa = 3.05EE485 pKa = 5.48FIVDD489 pKa = 3.7RR490 pKa = 11.84TDD492 pKa = 3.78DD493 pKa = 3.57VVNRR497 pKa = 11.84ISAVQDD503 pKa = 3.19EE504 pKa = 4.49PQFYY508 pKa = 10.17FDD510 pKa = 5.37CFGHH514 pKa = 6.93LKK516 pKa = 10.17RR517 pKa = 11.84ARR519 pKa = 11.84QMPVYY524 pKa = 8.72STPGLITRR532 pKa = 11.84LL533 pKa = 3.6
MM1 pKa = 7.27HH2 pKa = 7.49ASNYY6 pKa = 9.99KK7 pKa = 8.94MPSNMTHH14 pKa = 5.99NFSRR18 pKa = 11.84VPQVHH23 pKa = 5.96NPRR26 pKa = 11.84STFVSPYY33 pKa = 8.78TYY35 pKa = 9.28KK36 pKa = 10.25TALNAGFLYY45 pKa = 10.06PFFMDD50 pKa = 3.71EE51 pKa = 4.4VLPGDD56 pKa = 3.99TFNDD60 pKa = 3.61DD61 pKa = 3.38FTVFARR67 pKa = 11.84LTTPIKK73 pKa = 10.43PIMDD77 pKa = 4.32NLYY80 pKa = 10.07LDD82 pKa = 3.56TFFFYY87 pKa = 10.39IPNRR91 pKa = 11.84LLDD94 pKa = 4.11DD95 pKa = 3.29TWEE98 pKa = 4.35KK99 pKa = 11.43LNGAQDD105 pKa = 3.75NPGDD109 pKa = 4.17SIDD112 pKa = 3.84YY113 pKa = 9.17VVPQIPAPAVTGFASEE129 pKa = 4.43SLFDD133 pKa = 3.83YY134 pKa = 10.35MGIRR138 pKa = 11.84PGVTNFSIDD147 pKa = 3.18AYY149 pKa = 10.98HH150 pKa = 6.88PLAYY154 pKa = 10.57NLLFNTNFRR163 pKa = 11.84DD164 pKa = 3.47EE165 pKa = 4.24NLQDD169 pKa = 3.11SVYY172 pKa = 10.06FSKK175 pKa = 11.24GPGPHH180 pKa = 6.98AATNFNLLRR189 pKa = 11.84RR190 pKa = 11.84GKK192 pKa = 10.18RR193 pKa = 11.84FDD195 pKa = 3.87YY196 pKa = 8.15FTSCLPFPQKK206 pKa = 10.69GPEE209 pKa = 3.76VTIPILGDD217 pKa = 3.34APLIGIGTKK226 pKa = 9.98TNVGFSPGVTGYY238 pKa = 10.75DD239 pKa = 3.14YY240 pKa = 11.43DD241 pKa = 3.91GAVTYY246 pKa = 8.07TQGRR250 pKa = 11.84ATDD253 pKa = 3.58AGGANNAIIVDD264 pKa = 3.87VDD266 pKa = 3.71PATGLPKK273 pKa = 9.73WAASMAEE280 pKa = 3.91VSGVSINQFRR290 pKa = 11.84TSYY293 pKa = 9.7QIQMLLEE300 pKa = 4.04NDD302 pKa = 3.13ARR304 pKa = 11.84GGTRR308 pKa = 11.84YY309 pKa = 9.96PEE311 pKa = 3.8ILRR314 pKa = 11.84AHH316 pKa = 6.7FNVISPDD323 pKa = 3.24FRR325 pKa = 11.84LQRR328 pKa = 11.84PEE330 pKa = 3.9YY331 pKa = 10.47LGGTSVPVTVTPVPQTSSTDD351 pKa = 3.27NTSPQGNLAAYY362 pKa = 7.08ATSLNRR368 pKa = 11.84CGYY371 pKa = 7.33TKK373 pKa = 10.66SFVEE377 pKa = 4.15HH378 pKa = 6.35GVILGLCSIRR388 pKa = 11.84ADD390 pKa = 3.38LTYY393 pKa = 10.61QYY395 pKa = 11.07GLRR398 pKa = 11.84RR399 pKa = 11.84MWSRR403 pKa = 11.84QTRR406 pKa = 11.84YY407 pKa = 10.68DD408 pKa = 3.82FYY410 pKa = 10.82WPAFAHH416 pKa = 5.97LGEE419 pKa = 4.15MAVLNKK425 pKa = 10.06EE426 pKa = 4.09IYY428 pKa = 9.82IQGTADD434 pKa = 3.5DD435 pKa = 4.53NLVFGYY441 pKa = 10.16QEE443 pKa = 3.16AWADD447 pKa = 3.88YY448 pKa = 9.56KK449 pKa = 10.59YY450 pKa = 10.56HH451 pKa = 7.31PSLITGKK458 pKa = 8.75MRR460 pKa = 11.84SQDD463 pKa = 3.81PQSLDD468 pKa = 2.74IWHH471 pKa = 7.5LSQEE475 pKa = 4.43FQSLPLLNDD484 pKa = 3.05EE485 pKa = 5.48FIVDD489 pKa = 3.7RR490 pKa = 11.84TDD492 pKa = 3.78DD493 pKa = 3.57VVNRR497 pKa = 11.84ISAVQDD503 pKa = 3.19EE504 pKa = 4.49PQFYY508 pKa = 10.17FDD510 pKa = 5.37CFGHH514 pKa = 6.93LKK516 pKa = 10.17RR517 pKa = 11.84ARR519 pKa = 11.84QMPVYY524 pKa = 8.72STPGLITRR532 pKa = 11.84LL533 pKa = 3.6
Molecular weight: 59.92 kDa
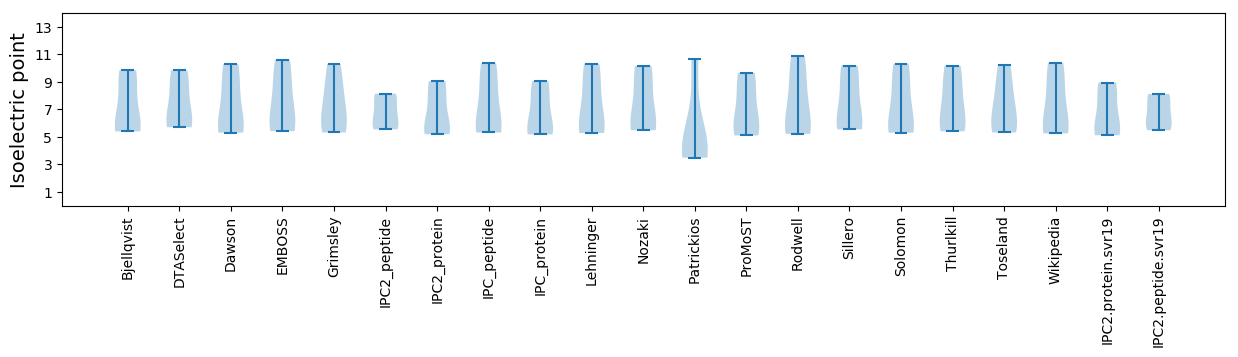
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UTX7|A0A3S8UTX7_9VIRU DNA pilot protein OS=Apis mellifera associated microvirus 9 OX=2494796 PE=4 SV=1
MM1 pKa = 7.56GLLSSIGKK9 pKa = 9.73AVGSITGGDD18 pKa = 3.45LLGFGGTLATGLLSNAGASAAQNFSAAQTQSQMDD52 pKa = 3.67FQEE55 pKa = 4.45RR56 pKa = 11.84MSNTAHH62 pKa = 5.61QRR64 pKa = 11.84QVADD68 pKa = 4.09LRR70 pKa = 11.84AAGLNPLLSATYY82 pKa = 9.67GGSSTPSGASAVGIDD97 pKa = 4.63KK98 pKa = 8.96ITPALASARR107 pKa = 11.84EE108 pKa = 4.09SARR111 pKa = 11.84VNAEE115 pKa = 4.2LKK117 pKa = 10.89NMQATNSKK125 pKa = 9.7IDD127 pKa = 3.08SDD129 pKa = 3.92TRR131 pKa = 11.84LNNALTVSAKK141 pKa = 10.61ADD143 pKa = 3.83ALSKK147 pKa = 10.4AASARR152 pKa = 11.84ASNAAADD159 pKa = 4.06TTLAMLPGRR168 pKa = 11.84EE169 pKa = 4.03AQSKK173 pKa = 10.48NSAKK177 pKa = 9.67EE178 pKa = 3.94AKK180 pKa = 10.15SFWATHH186 pKa = 4.47IRR188 pKa = 11.84TRR190 pKa = 11.84TKK192 pKa = 10.56PFFEE196 pKa = 5.13SIGDD200 pKa = 3.5ATGAVGNLFSGSKK213 pKa = 9.52QDD215 pKa = 3.19SGSMSRR221 pKa = 11.84FLSGGLL227 pKa = 3.47
MM1 pKa = 7.56GLLSSIGKK9 pKa = 9.73AVGSITGGDD18 pKa = 3.45LLGFGGTLATGLLSNAGASAAQNFSAAQTQSQMDD52 pKa = 3.67FQEE55 pKa = 4.45RR56 pKa = 11.84MSNTAHH62 pKa = 5.61QRR64 pKa = 11.84QVADD68 pKa = 4.09LRR70 pKa = 11.84AAGLNPLLSATYY82 pKa = 9.67GGSSTPSGASAVGIDD97 pKa = 4.63KK98 pKa = 8.96ITPALASARR107 pKa = 11.84EE108 pKa = 4.09SARR111 pKa = 11.84VNAEE115 pKa = 4.2LKK117 pKa = 10.89NMQATNSKK125 pKa = 9.7IDD127 pKa = 3.08SDD129 pKa = 3.92TRR131 pKa = 11.84LNNALTVSAKK141 pKa = 10.61ADD143 pKa = 3.83ALSKK147 pKa = 10.4AASARR152 pKa = 11.84ASNAAADD159 pKa = 4.06TTLAMLPGRR168 pKa = 11.84EE169 pKa = 4.03AQSKK173 pKa = 10.48NSAKK177 pKa = 9.67EE178 pKa = 3.94AKK180 pKa = 10.15SFWATHH186 pKa = 4.47IRR188 pKa = 11.84TRR190 pKa = 11.84TKK192 pKa = 10.56PFFEE196 pKa = 5.13SIGDD200 pKa = 3.5ATGAVGNLFSGSKK213 pKa = 9.52QDD215 pKa = 3.19SGSMSRR221 pKa = 11.84FLSGGLL227 pKa = 3.47
Molecular weight: 23.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1352 |
87 |
533 |
270.4 |
30.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.876 ± 1.608 | 1.109 ± 0.634 |
6.731 ± 0.425 | 3.846 ± 0.66 |
5.251 ± 0.597 | 7.544 ± 0.63 |
2.441 ± 0.414 | 3.846 ± 0.47 |
4.29 ± 0.834 | 7.766 ± 0.489 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.997 ± 0.225 | 4.808 ± 0.501 |
5.991 ± 1.145 | 3.624 ± 0.562 |
6.361 ± 0.832 | 7.988 ± 1.142 |
6.509 ± 0.593 | 5.547 ± 0.566 |
0.962 ± 0.232 | 4.512 ± 0.945 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
