
Mytilus sp. clam associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.68
Get precalculated fractions of proteins
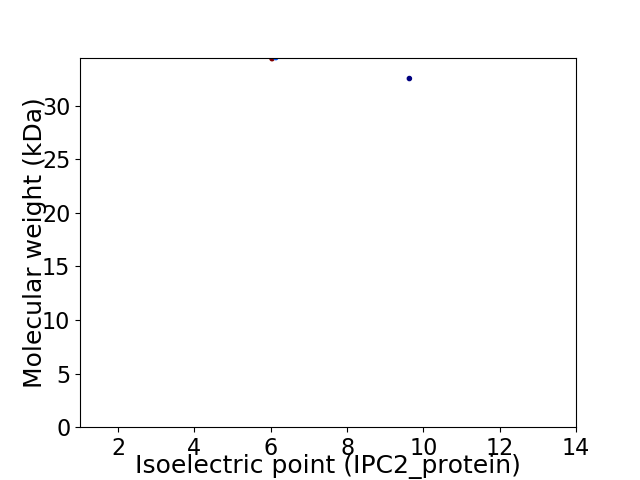
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
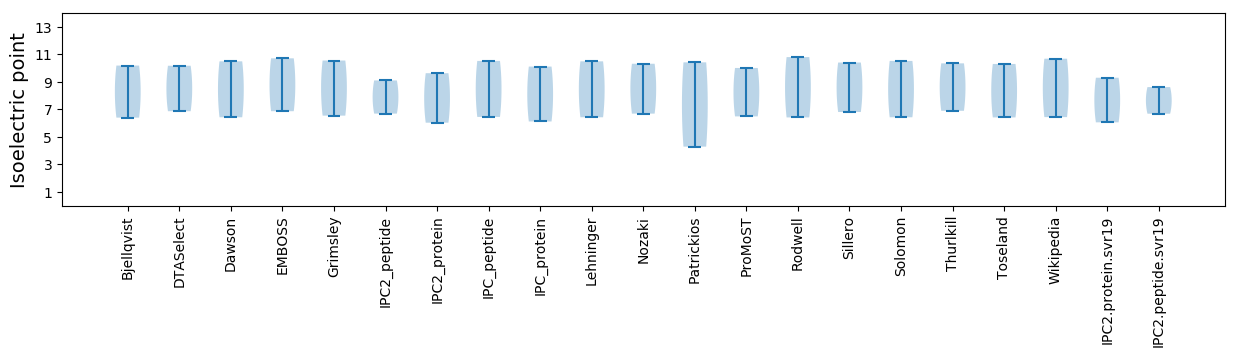
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1RL85|A0A0K1RL85_9CIRC Putative replication initiation protein OS=Mytilus sp. clam associated circular virus OX=1692256 PE=4 SV=1
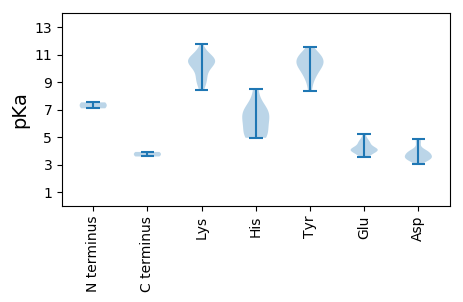
MM1 pKa = 7.09SQAKK5 pKa = 9.28RR6 pKa = 11.84WCFTINNPTPAEE18 pKa = 4.03EE19 pKa = 4.14EE20 pKa = 4.49SLFLHH25 pKa = 6.61GTNIDD30 pKa = 3.97HH31 pKa = 7.24PPCDD35 pKa = 3.52FKK37 pKa = 11.76YY38 pKa = 10.65LVFGRR43 pKa = 11.84EE44 pKa = 4.05TGEE47 pKa = 3.73SGTPHH52 pKa = 5.65LQGYY56 pKa = 8.86FEE58 pKa = 4.63LVKK61 pKa = 10.49KK62 pKa = 10.62LRR64 pKa = 11.84IHH66 pKa = 6.03QIKK69 pKa = 8.42GTLGFEE75 pKa = 4.13RR76 pKa = 11.84SHH78 pKa = 8.51LEE80 pKa = 3.88VARR83 pKa = 11.84GTSLQASDD91 pKa = 3.71YY92 pKa = 10.09CKK94 pKa = 10.61KK95 pKa = 10.48EE96 pKa = 3.61NAFEE100 pKa = 4.14EE101 pKa = 5.23FGALPPPPGNAAHH114 pKa = 6.91FAQLRR119 pKa = 11.84EE120 pKa = 4.18WVAAQPSKK128 pKa = 10.04PTIKK132 pKa = 10.54DD133 pKa = 3.06VWDD136 pKa = 3.69VFPTLAARR144 pKa = 11.84YY145 pKa = 8.34NRR147 pKa = 11.84AVMEE151 pKa = 4.96CIDD154 pKa = 4.59LFGKK158 pKa = 10.56KK159 pKa = 9.1PVLVEE164 pKa = 3.58GDD166 pKa = 3.31LRR168 pKa = 11.84LWQLRR173 pKa = 11.84LDD175 pKa = 4.83GIANQEE181 pKa = 3.65ADD183 pKa = 3.63DD184 pKa = 3.91RR185 pKa = 11.84RR186 pKa = 11.84VIFVIDD192 pKa = 3.55PEE194 pKa = 4.61GNKK197 pKa = 9.95GKK199 pKa = 10.11SWLVSYY205 pKa = 9.59WLSTRR210 pKa = 11.84DD211 pKa = 3.29DD212 pKa = 3.88TQFMSVGKK220 pKa = 10.22RR221 pKa = 11.84DD222 pKa = 3.82DD223 pKa = 3.2LAYY226 pKa = 10.34AVDD229 pKa = 4.03ISTCLFVFDD238 pKa = 4.47IPRR241 pKa = 11.84GNMQYY246 pKa = 9.32IQYY249 pKa = 11.03GIFEE253 pKa = 4.15QLKK256 pKa = 8.84NRR258 pKa = 11.84VVFSNKK264 pKa = 9.43YY265 pKa = 9.35SSQTKK270 pKa = 9.71ILLNTPHH277 pKa = 6.41VFVFANEE284 pKa = 4.06LPDD287 pKa = 3.86MNALSADD294 pKa = 3.83RR295 pKa = 11.84YY296 pKa = 10.46KK297 pKa = 11.09VINII301 pKa = 3.92
MM1 pKa = 7.09SQAKK5 pKa = 9.28RR6 pKa = 11.84WCFTINNPTPAEE18 pKa = 4.03EE19 pKa = 4.14EE20 pKa = 4.49SLFLHH25 pKa = 6.61GTNIDD30 pKa = 3.97HH31 pKa = 7.24PPCDD35 pKa = 3.52FKK37 pKa = 11.76YY38 pKa = 10.65LVFGRR43 pKa = 11.84EE44 pKa = 4.05TGEE47 pKa = 3.73SGTPHH52 pKa = 5.65LQGYY56 pKa = 8.86FEE58 pKa = 4.63LVKK61 pKa = 10.49KK62 pKa = 10.62LRR64 pKa = 11.84IHH66 pKa = 6.03QIKK69 pKa = 8.42GTLGFEE75 pKa = 4.13RR76 pKa = 11.84SHH78 pKa = 8.51LEE80 pKa = 3.88VARR83 pKa = 11.84GTSLQASDD91 pKa = 3.71YY92 pKa = 10.09CKK94 pKa = 10.61KK95 pKa = 10.48EE96 pKa = 3.61NAFEE100 pKa = 4.14EE101 pKa = 5.23FGALPPPPGNAAHH114 pKa = 6.91FAQLRR119 pKa = 11.84EE120 pKa = 4.18WVAAQPSKK128 pKa = 10.04PTIKK132 pKa = 10.54DD133 pKa = 3.06VWDD136 pKa = 3.69VFPTLAARR144 pKa = 11.84YY145 pKa = 8.34NRR147 pKa = 11.84AVMEE151 pKa = 4.96CIDD154 pKa = 4.59LFGKK158 pKa = 10.56KK159 pKa = 9.1PVLVEE164 pKa = 3.58GDD166 pKa = 3.31LRR168 pKa = 11.84LWQLRR173 pKa = 11.84LDD175 pKa = 4.83GIANQEE181 pKa = 3.65ADD183 pKa = 3.63DD184 pKa = 3.91RR185 pKa = 11.84RR186 pKa = 11.84VIFVIDD192 pKa = 3.55PEE194 pKa = 4.61GNKK197 pKa = 9.95GKK199 pKa = 10.11SWLVSYY205 pKa = 9.59WLSTRR210 pKa = 11.84DD211 pKa = 3.29DD212 pKa = 3.88TQFMSVGKK220 pKa = 10.22RR221 pKa = 11.84DD222 pKa = 3.82DD223 pKa = 3.2LAYY226 pKa = 10.34AVDD229 pKa = 4.03ISTCLFVFDD238 pKa = 4.47IPRR241 pKa = 11.84GNMQYY246 pKa = 9.32IQYY249 pKa = 11.03GIFEE253 pKa = 4.15QLKK256 pKa = 8.84NRR258 pKa = 11.84VVFSNKK264 pKa = 9.43YY265 pKa = 9.35SSQTKK270 pKa = 9.71ILLNTPHH277 pKa = 6.41VFVFANEE284 pKa = 4.06LPDD287 pKa = 3.86MNALSADD294 pKa = 3.83RR295 pKa = 11.84YY296 pKa = 10.46KK297 pKa = 11.09VINII301 pKa = 3.92
Molecular weight: 34.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1RL85|A0A0K1RL85_9CIRC Putative replication initiation protein OS=Mytilus sp. clam associated circular virus OX=1692256 PE=4 SV=1
MM1 pKa = 7.52SGQLVRR7 pKa = 11.84SFARR11 pKa = 11.84AQYY14 pKa = 10.52PRR16 pKa = 11.84FRR18 pKa = 11.84NYY20 pKa = 10.81AATRR24 pKa = 11.84LQAAGRR30 pKa = 11.84GWLSRR35 pKa = 11.84RR36 pKa = 11.84TRR38 pKa = 11.84GAGQTFQRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84MWSGRR54 pKa = 11.84SRR56 pKa = 11.84KK57 pKa = 8.5RR58 pKa = 11.84TGQMRR63 pKa = 11.84KK64 pKa = 9.04RR65 pKa = 11.84MAVGTRR71 pKa = 11.84IGSSLAKK78 pKa = 9.99CDD80 pKa = 3.53QVEE83 pKa = 4.72DD84 pKa = 4.01NFSATTKK91 pKa = 10.37EE92 pKa = 3.9LYY94 pKa = 9.76QYY96 pKa = 11.56KK97 pKa = 10.58LLDD100 pKa = 3.59LAKK103 pKa = 10.81NNNADD108 pKa = 3.54DD109 pKa = 4.03TQVRR113 pKa = 11.84NRR115 pKa = 11.84DD116 pKa = 3.34VVNFRR121 pKa = 11.84GIKK124 pKa = 10.04LCINTTNTTTTGTIYY139 pKa = 10.46PNRR142 pKa = 11.84HH143 pKa = 4.95LHH145 pKa = 4.99CHH147 pKa = 5.19YY148 pKa = 11.06AVVSPKK154 pKa = 10.85ALDD157 pKa = 3.34ATQAVPYY164 pKa = 9.92GHH166 pKa = 7.13FFRR169 pKa = 11.84SQGEE173 pKa = 4.04VGKK176 pKa = 10.37RR177 pKa = 11.84GKK179 pKa = 10.48DD180 pKa = 3.26FGVDD184 pKa = 3.57LNGMDD189 pKa = 4.81FQCLPINTDD198 pKa = 3.47LYY200 pKa = 10.7TIHH203 pKa = 5.12QHH205 pKa = 6.43YY206 pKa = 10.22RR207 pKa = 11.84FKK209 pKa = 11.15LNKK212 pKa = 9.09AANSSTSPEE221 pKa = 3.93KK222 pKa = 10.76LYY224 pKa = 11.07EE225 pKa = 4.42KK226 pKa = 10.56YY227 pKa = 10.99LNLNRR232 pKa = 11.84QVRR235 pKa = 11.84YY236 pKa = 9.99EE237 pKa = 3.99NTGDD241 pKa = 3.48KK242 pKa = 11.02PEE244 pKa = 4.23GKK246 pKa = 9.52NVYY249 pKa = 10.25LLIWFSFIDD258 pKa = 3.98EE259 pKa = 4.68AGGTAGIPCADD270 pKa = 3.46TKK272 pKa = 11.28LRR274 pKa = 11.84ITRR277 pKa = 11.84FFKK280 pKa = 10.68EE281 pKa = 4.07SSNN284 pKa = 3.58
MM1 pKa = 7.52SGQLVRR7 pKa = 11.84SFARR11 pKa = 11.84AQYY14 pKa = 10.52PRR16 pKa = 11.84FRR18 pKa = 11.84NYY20 pKa = 10.81AATRR24 pKa = 11.84LQAAGRR30 pKa = 11.84GWLSRR35 pKa = 11.84RR36 pKa = 11.84TRR38 pKa = 11.84GAGQTFQRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84MWSGRR54 pKa = 11.84SRR56 pKa = 11.84KK57 pKa = 8.5RR58 pKa = 11.84TGQMRR63 pKa = 11.84KK64 pKa = 9.04RR65 pKa = 11.84MAVGTRR71 pKa = 11.84IGSSLAKK78 pKa = 9.99CDD80 pKa = 3.53QVEE83 pKa = 4.72DD84 pKa = 4.01NFSATTKK91 pKa = 10.37EE92 pKa = 3.9LYY94 pKa = 9.76QYY96 pKa = 11.56KK97 pKa = 10.58LLDD100 pKa = 3.59LAKK103 pKa = 10.81NNNADD108 pKa = 3.54DD109 pKa = 4.03TQVRR113 pKa = 11.84NRR115 pKa = 11.84DD116 pKa = 3.34VVNFRR121 pKa = 11.84GIKK124 pKa = 10.04LCINTTNTTTTGTIYY139 pKa = 10.46PNRR142 pKa = 11.84HH143 pKa = 4.95LHH145 pKa = 4.99CHH147 pKa = 5.19YY148 pKa = 11.06AVVSPKK154 pKa = 10.85ALDD157 pKa = 3.34ATQAVPYY164 pKa = 9.92GHH166 pKa = 7.13FFRR169 pKa = 11.84SQGEE173 pKa = 4.04VGKK176 pKa = 10.37RR177 pKa = 11.84GKK179 pKa = 10.48DD180 pKa = 3.26FGVDD184 pKa = 3.57LNGMDD189 pKa = 4.81FQCLPINTDD198 pKa = 3.47LYY200 pKa = 10.7TIHH203 pKa = 5.12QHH205 pKa = 6.43YY206 pKa = 10.22RR207 pKa = 11.84FKK209 pKa = 11.15LNKK212 pKa = 9.09AANSSTSPEE221 pKa = 3.93KK222 pKa = 10.76LYY224 pKa = 11.07EE225 pKa = 4.42KK226 pKa = 10.56YY227 pKa = 10.99LNLNRR232 pKa = 11.84QVRR235 pKa = 11.84YY236 pKa = 9.99EE237 pKa = 3.99NTGDD241 pKa = 3.48KK242 pKa = 11.02PEE244 pKa = 4.23GKK246 pKa = 9.52NVYY249 pKa = 10.25LLIWFSFIDD258 pKa = 3.98EE259 pKa = 4.68AGGTAGIPCADD270 pKa = 3.46TKK272 pKa = 11.28LRR274 pKa = 11.84ITRR277 pKa = 11.84FFKK280 pKa = 10.68EE281 pKa = 4.07SSNN284 pKa = 3.58
Molecular weight: 32.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
585 |
284 |
301 |
292.5 |
33.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.179 ± 0.149 | 1.709 ± 0.035 |
5.641 ± 0.493 | 4.615 ± 1.002 |
5.641 ± 0.493 | 6.838 ± 0.63 |
2.222 ± 0.076 | 4.444 ± 0.64 |
6.154 ± 0.128 | 8.205 ± 0.562 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.709 ± 0.035 | 5.812 ± 0.609 |
4.274 ± 1.009 | 4.615 ± 0.218 |
8.034 ± 1.753 | 5.47 ± 0.113 |
6.325 ± 1.229 | 5.641 ± 0.737 |
1.538 ± 0.334 | 3.932 ± 0.448 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
