
Myxococcus xanthus (strain DK1622)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Cystobacterineae; Myxococcaceae; Myxococcus; Myxococcus xanthus
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
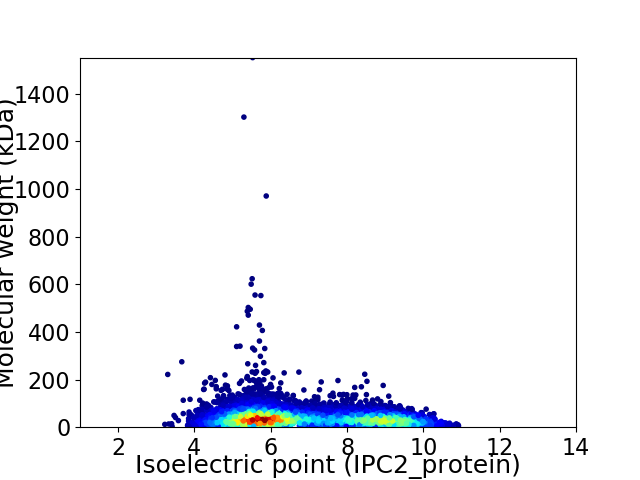
Virtual 2D-PAGE plot for 7314 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q1CYQ9|Q1CYQ9_MYXXD Vitamin K epoxide reductase family/thioredoxin domain protein OS=Myxococcus xanthus (strain DK1622) OX=246197 GN=MXAN_6339 PE=3 SV=1
MM1 pKa = 7.34EE2 pKa = 4.2QTAHH6 pKa = 6.35MGTNRR11 pKa = 11.84QTWLALTLTVVGLLAGCSPGASPTEE36 pKa = 3.87EE37 pKa = 4.72DD38 pKa = 3.51ADD40 pKa = 4.08GDD42 pKa = 4.42GVADD46 pKa = 4.92AADD49 pKa = 4.46CAPGDD54 pKa = 3.82ANRR57 pKa = 11.84WRR59 pKa = 11.84ILEE62 pKa = 3.87GHH64 pKa = 7.09ADD66 pKa = 3.83DD67 pKa = 6.17DD68 pKa = 4.79RR69 pKa = 11.84DD70 pKa = 4.01GVGAGALRR78 pKa = 11.84EE79 pKa = 4.26VCAGEE84 pKa = 3.86QLPEE88 pKa = 4.01GWSQQSGDD96 pKa = 4.52CAPGDD101 pKa = 3.46ATRR104 pKa = 11.84WRR106 pKa = 11.84EE107 pKa = 3.89QAGLYY112 pKa = 9.54PDD114 pKa = 3.94QDD116 pKa = 3.44NDD118 pKa = 3.61GATGEE123 pKa = 4.58GPVTACVGDD132 pKa = 3.67TLEE135 pKa = 5.28GYY137 pKa = 9.79RR138 pKa = 11.84EE139 pKa = 4.02QPGAPDD145 pKa = 4.41CDD147 pKa = 4.18DD148 pKa = 3.65NAPRR152 pKa = 11.84FAASTQLWPDD162 pKa = 3.07ADD164 pKa = 3.89FDD166 pKa = 5.14GIGNGEE172 pKa = 4.68PISHH176 pKa = 7.07CAGKK180 pKa = 10.1DD181 pKa = 3.4LPRR184 pKa = 11.84GYY186 pKa = 10.83AAVDD190 pKa = 3.92GDD192 pKa = 4.41CAPDD196 pKa = 3.39DD197 pKa = 3.95RR198 pKa = 11.84ARR200 pKa = 11.84STWRR204 pKa = 11.84DD205 pKa = 3.36YY206 pKa = 7.45THH208 pKa = 7.57RR209 pKa = 11.84DD210 pKa = 2.94ADD212 pKa = 3.77GDD214 pKa = 4.01GFTVPEE220 pKa = 4.29EE221 pKa = 4.16GRR223 pKa = 11.84VCSGMTLPAGYY234 pKa = 10.38ALEE237 pKa = 4.95ARR239 pKa = 11.84GLDD242 pKa = 4.15CDD244 pKa = 4.36DD245 pKa = 4.87TNAAWWQKK253 pKa = 7.72RR254 pKa = 11.84TVYY257 pKa = 11.17ADD259 pKa = 3.1VDD261 pKa = 3.7GDD263 pKa = 4.2GFGSGDD269 pKa = 3.12PVMRR273 pKa = 11.84CTGRR277 pKa = 11.84TAPPGYY283 pKa = 10.7ALDD286 pKa = 4.46SVDD289 pKa = 5.18CAPEE293 pKa = 4.64DD294 pKa = 3.37ATRR297 pKa = 11.84WQWRR301 pKa = 11.84SYY303 pKa = 9.37AHH305 pKa = 7.25RR306 pKa = 11.84DD307 pKa = 3.13SDD309 pKa = 4.18GDD311 pKa = 3.7GDD313 pKa = 4.22TVPEE317 pKa = 4.08QGVTCSGAALPPGYY331 pKa = 10.2AQWPTGHH338 pKa = 7.33DD339 pKa = 3.81CNDD342 pKa = 3.22RR343 pKa = 11.84DD344 pKa = 3.96ATVMVTWHH352 pKa = 6.51VYY354 pKa = 10.83SDD356 pKa = 3.96DD357 pKa = 4.24DD358 pKa = 4.46GDD360 pKa = 3.86RR361 pKa = 11.84FGAGTRR367 pKa = 11.84QTFCAGRR374 pKa = 11.84QIPVGYY380 pKa = 10.02SVVNTDD386 pKa = 4.89CDD388 pKa = 3.7DD389 pKa = 3.81TAPLLWRR396 pKa = 11.84LLAYY400 pKa = 8.0SHH402 pKa = 7.27RR403 pKa = 11.84DD404 pKa = 3.01ADD406 pKa = 4.03GDD408 pKa = 4.12SFTAPEE414 pKa = 4.19VGTVCSGMALPPGHH428 pKa = 6.71ATTPRR433 pKa = 11.84GNDD436 pKa = 3.69CDD438 pKa = 4.63DD439 pKa = 4.86ADD441 pKa = 4.18PSIHH445 pKa = 5.67TTVQAWTDD453 pKa = 3.22ADD455 pKa = 3.94GDD457 pKa = 4.41GVGAGDD463 pKa = 4.94AVALCTDD470 pKa = 4.09GTVPAPWSARR480 pKa = 11.84GTDD483 pKa = 3.94CAAEE487 pKa = 4.08DD488 pKa = 3.49AARR491 pKa = 11.84WRR493 pKa = 11.84QAAYY497 pKa = 10.53AHH499 pKa = 6.48VDD501 pKa = 3.28RR502 pKa = 11.84DD503 pKa = 4.15ADD505 pKa = 3.74GHH507 pKa = 4.83THH509 pKa = 6.93PEE511 pKa = 3.93AGTVCTGASLPEE523 pKa = 4.29PYY525 pKa = 8.66FTQATGNDD533 pKa = 4.12CDD535 pKa = 5.23DD536 pKa = 3.95SDD538 pKa = 3.72AARR541 pKa = 11.84YY542 pKa = 8.31RR543 pKa = 11.84WTVLYY548 pKa = 10.17PDD550 pKa = 3.89MDD552 pKa = 4.73GDD554 pKa = 4.72GVGTSPRR561 pKa = 11.84TIPCIGAQTPLGFSLKK577 pKa = 10.49GYY579 pKa = 10.6DD580 pKa = 4.36VDD582 pKa = 6.27DD583 pKa = 4.39SDD585 pKa = 6.46ASIQEE590 pKa = 4.24DD591 pKa = 4.26PEE593 pKa = 6.58DD594 pKa = 4.52DD595 pKa = 4.87LMLEE599 pKa = 5.14LILDD603 pKa = 4.07
MM1 pKa = 7.34EE2 pKa = 4.2QTAHH6 pKa = 6.35MGTNRR11 pKa = 11.84QTWLALTLTVVGLLAGCSPGASPTEE36 pKa = 3.87EE37 pKa = 4.72DD38 pKa = 3.51ADD40 pKa = 4.08GDD42 pKa = 4.42GVADD46 pKa = 4.92AADD49 pKa = 4.46CAPGDD54 pKa = 3.82ANRR57 pKa = 11.84WRR59 pKa = 11.84ILEE62 pKa = 3.87GHH64 pKa = 7.09ADD66 pKa = 3.83DD67 pKa = 6.17DD68 pKa = 4.79RR69 pKa = 11.84DD70 pKa = 4.01GVGAGALRR78 pKa = 11.84EE79 pKa = 4.26VCAGEE84 pKa = 3.86QLPEE88 pKa = 4.01GWSQQSGDD96 pKa = 4.52CAPGDD101 pKa = 3.46ATRR104 pKa = 11.84WRR106 pKa = 11.84EE107 pKa = 3.89QAGLYY112 pKa = 9.54PDD114 pKa = 3.94QDD116 pKa = 3.44NDD118 pKa = 3.61GATGEE123 pKa = 4.58GPVTACVGDD132 pKa = 3.67TLEE135 pKa = 5.28GYY137 pKa = 9.79RR138 pKa = 11.84EE139 pKa = 4.02QPGAPDD145 pKa = 4.41CDD147 pKa = 4.18DD148 pKa = 3.65NAPRR152 pKa = 11.84FAASTQLWPDD162 pKa = 3.07ADD164 pKa = 3.89FDD166 pKa = 5.14GIGNGEE172 pKa = 4.68PISHH176 pKa = 7.07CAGKK180 pKa = 10.1DD181 pKa = 3.4LPRR184 pKa = 11.84GYY186 pKa = 10.83AAVDD190 pKa = 3.92GDD192 pKa = 4.41CAPDD196 pKa = 3.39DD197 pKa = 3.95RR198 pKa = 11.84ARR200 pKa = 11.84STWRR204 pKa = 11.84DD205 pKa = 3.36YY206 pKa = 7.45THH208 pKa = 7.57RR209 pKa = 11.84DD210 pKa = 2.94ADD212 pKa = 3.77GDD214 pKa = 4.01GFTVPEE220 pKa = 4.29EE221 pKa = 4.16GRR223 pKa = 11.84VCSGMTLPAGYY234 pKa = 10.38ALEE237 pKa = 4.95ARR239 pKa = 11.84GLDD242 pKa = 4.15CDD244 pKa = 4.36DD245 pKa = 4.87TNAAWWQKK253 pKa = 7.72RR254 pKa = 11.84TVYY257 pKa = 11.17ADD259 pKa = 3.1VDD261 pKa = 3.7GDD263 pKa = 4.2GFGSGDD269 pKa = 3.12PVMRR273 pKa = 11.84CTGRR277 pKa = 11.84TAPPGYY283 pKa = 10.7ALDD286 pKa = 4.46SVDD289 pKa = 5.18CAPEE293 pKa = 4.64DD294 pKa = 3.37ATRR297 pKa = 11.84WQWRR301 pKa = 11.84SYY303 pKa = 9.37AHH305 pKa = 7.25RR306 pKa = 11.84DD307 pKa = 3.13SDD309 pKa = 4.18GDD311 pKa = 3.7GDD313 pKa = 4.22TVPEE317 pKa = 4.08QGVTCSGAALPPGYY331 pKa = 10.2AQWPTGHH338 pKa = 7.33DD339 pKa = 3.81CNDD342 pKa = 3.22RR343 pKa = 11.84DD344 pKa = 3.96ATVMVTWHH352 pKa = 6.51VYY354 pKa = 10.83SDD356 pKa = 3.96DD357 pKa = 4.24DD358 pKa = 4.46GDD360 pKa = 3.86RR361 pKa = 11.84FGAGTRR367 pKa = 11.84QTFCAGRR374 pKa = 11.84QIPVGYY380 pKa = 10.02SVVNTDD386 pKa = 4.89CDD388 pKa = 3.7DD389 pKa = 3.81TAPLLWRR396 pKa = 11.84LLAYY400 pKa = 8.0SHH402 pKa = 7.27RR403 pKa = 11.84DD404 pKa = 3.01ADD406 pKa = 4.03GDD408 pKa = 4.12SFTAPEE414 pKa = 4.19VGTVCSGMALPPGHH428 pKa = 6.71ATTPRR433 pKa = 11.84GNDD436 pKa = 3.69CDD438 pKa = 4.63DD439 pKa = 4.86ADD441 pKa = 4.18PSIHH445 pKa = 5.67TTVQAWTDD453 pKa = 3.22ADD455 pKa = 3.94GDD457 pKa = 4.41GVGAGDD463 pKa = 4.94AVALCTDD470 pKa = 4.09GTVPAPWSARR480 pKa = 11.84GTDD483 pKa = 3.94CAAEE487 pKa = 4.08DD488 pKa = 3.49AARR491 pKa = 11.84WRR493 pKa = 11.84QAAYY497 pKa = 10.53AHH499 pKa = 6.48VDD501 pKa = 3.28RR502 pKa = 11.84DD503 pKa = 4.15ADD505 pKa = 3.74GHH507 pKa = 4.83THH509 pKa = 6.93PEE511 pKa = 3.93AGTVCTGASLPEE523 pKa = 4.29PYY525 pKa = 8.66FTQATGNDD533 pKa = 4.12CDD535 pKa = 5.23DD536 pKa = 3.95SDD538 pKa = 3.72AARR541 pKa = 11.84YY542 pKa = 8.31RR543 pKa = 11.84WTVLYY548 pKa = 10.17PDD550 pKa = 3.89MDD552 pKa = 4.73GDD554 pKa = 4.72GVGTSPRR561 pKa = 11.84TIPCIGAQTPLGFSLKK577 pKa = 10.49GYY579 pKa = 10.6DD580 pKa = 4.36VDD582 pKa = 6.27DD583 pKa = 4.39SDD585 pKa = 6.46ASIQEE590 pKa = 4.24DD591 pKa = 4.26PEE593 pKa = 6.58DD594 pKa = 4.52DD595 pKa = 4.87LMLEE599 pKa = 5.14LILDD603 pKa = 4.07
Molecular weight: 63.72 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q1CXK1|Q1CXK1_MYXXD Uncharacterized protein OS=Myxococcus xanthus (strain DK1622) OX=246197 GN=MXAN_6755 PE=4 SV=1
MM1 pKa = 7.34TNEE4 pKa = 4.26SVNPQLTDD12 pKa = 3.58DD13 pKa = 4.37VSPSPAEE20 pKa = 4.0AGIAPTPRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84TATKK34 pKa = 9.67RR35 pKa = 11.84KK36 pKa = 8.09SASRR40 pKa = 11.84TGSTRR45 pKa = 11.84KK46 pKa = 9.4SARR49 pKa = 11.84KK50 pKa = 8.2ATAKK54 pKa = 10.4RR55 pKa = 11.84GTPRR59 pKa = 11.84AKK61 pKa = 9.24KK62 pKa = 7.44TAARR66 pKa = 11.84GKK68 pKa = 7.71TATRR72 pKa = 11.84KK73 pKa = 9.32SATRR77 pKa = 11.84KK78 pKa = 8.68SATRR82 pKa = 11.84ATGRR86 pKa = 11.84GARR89 pKa = 11.84KK90 pKa = 7.88ATPRR94 pKa = 11.84KK95 pKa = 7.83TGTAGRR101 pKa = 11.84TARR104 pKa = 11.84KK105 pKa = 7.6TGTRR109 pKa = 11.84GTATKK114 pKa = 9.5RR115 pKa = 11.84TTTGGGARR123 pKa = 11.84KK124 pKa = 9.78SPARR128 pKa = 11.84RR129 pKa = 11.84TTRR132 pKa = 11.84RR133 pKa = 3.17
MM1 pKa = 7.34TNEE4 pKa = 4.26SVNPQLTDD12 pKa = 3.58DD13 pKa = 4.37VSPSPAEE20 pKa = 4.0AGIAPTPRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84TATKK34 pKa = 9.67RR35 pKa = 11.84KK36 pKa = 8.09SASRR40 pKa = 11.84TGSTRR45 pKa = 11.84KK46 pKa = 9.4SARR49 pKa = 11.84KK50 pKa = 8.2ATAKK54 pKa = 10.4RR55 pKa = 11.84GTPRR59 pKa = 11.84AKK61 pKa = 9.24KK62 pKa = 7.44TAARR66 pKa = 11.84GKK68 pKa = 7.71TATRR72 pKa = 11.84KK73 pKa = 9.32SATRR77 pKa = 11.84KK78 pKa = 8.68SATRR82 pKa = 11.84ATGRR86 pKa = 11.84GARR89 pKa = 11.84KK90 pKa = 7.88ATPRR94 pKa = 11.84KK95 pKa = 7.83TGTAGRR101 pKa = 11.84TARR104 pKa = 11.84KK105 pKa = 7.6TGTRR109 pKa = 11.84GTATKK114 pKa = 9.5RR115 pKa = 11.84TTTGGGARR123 pKa = 11.84KK124 pKa = 9.78SPARR128 pKa = 11.84RR129 pKa = 11.84TTRR132 pKa = 11.84RR133 pKa = 3.17
Molecular weight: 14.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2775111 |
30 |
14274 |
379.4 |
41.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.228 ± 0.045 | 0.95 ± 0.013 |
5.119 ± 0.02 | 6.339 ± 0.032 |
3.305 ± 0.016 | 8.711 ± 0.03 |
2.178 ± 0.015 | 2.914 ± 0.022 |
2.81 ± 0.031 | 10.911 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.026 ± 0.013 | 2.098 ± 0.018 |
6.173 ± 0.027 | 3.427 ± 0.02 |
8.168 ± 0.032 | 5.688 ± 0.026 |
5.367 ± 0.026 | 8.209 ± 0.03 |
1.362 ± 0.012 | 2.019 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
