
Beihai picorna-like virus 118
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
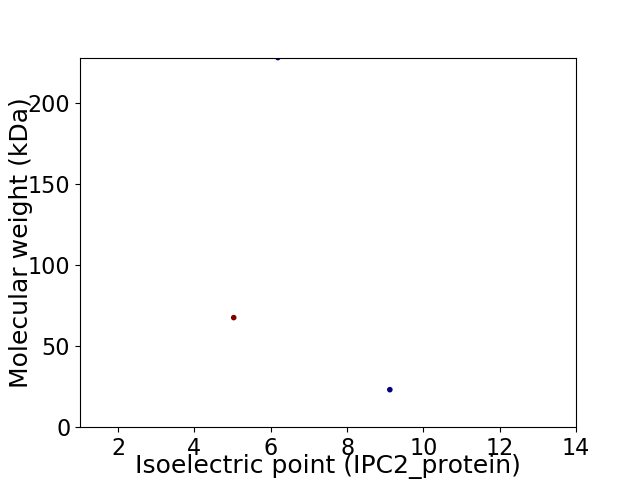
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KH81|A0A1L3KH81_9VIRU Uncharacterized protein OS=Beihai picorna-like virus 118 OX=1922547 PE=4 SV=1
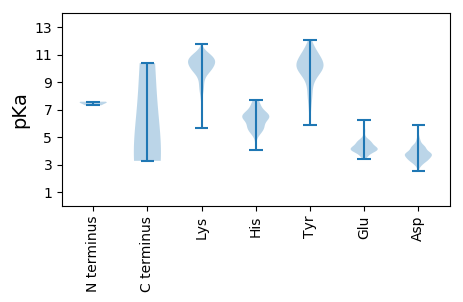
MM1 pKa = 7.57SDD3 pKa = 4.1KK4 pKa = 10.8DD5 pKa = 3.42AAMPPTASGEE15 pKa = 4.04ITEE18 pKa = 4.66GMGVPPEE25 pKa = 4.13MPVDD29 pKa = 3.85AGIDD33 pKa = 3.46RR34 pKa = 11.84PGGITAEE41 pKa = 4.24VQQVHH46 pKa = 5.63TSRR49 pKa = 11.84NDD51 pKa = 3.34SIEE54 pKa = 3.88AFLRR58 pKa = 11.84SVPILQKK65 pKa = 10.13IIKK68 pKa = 9.18WSVNDD73 pKa = 3.67LPGTILYY80 pKa = 7.77QVPVHH85 pKa = 6.31PSYY88 pKa = 11.46VNYY91 pKa = 8.4MVKK94 pKa = 9.67WFKK97 pKa = 10.83KK98 pKa = 9.74AFKK101 pKa = 9.25YY102 pKa = 9.99WNGPIIFMIDD112 pKa = 3.04LAGTGFHH119 pKa = 6.53GGKK122 pKa = 10.26LIIFHH127 pKa = 6.4TPPGKK132 pKa = 10.03RR133 pKa = 11.84FKK135 pKa = 10.0PSKK138 pKa = 8.35MTMAQWFTFSHH149 pKa = 6.4GLVDD153 pKa = 5.84AKK155 pKa = 11.22SQGNNLACAPDD166 pKa = 3.86VIGRR170 pKa = 11.84TLHH173 pKa = 7.23DD174 pKa = 4.26NGPFDD179 pKa = 4.78EE180 pKa = 4.87NNEE183 pKa = 3.99DD184 pKa = 4.15TYY186 pKa = 11.73GGWFGIGVVMEE197 pKa = 5.1LKK199 pKa = 10.16TSGTGDD205 pKa = 3.36QEE207 pKa = 4.17IQFSLEE213 pKa = 3.57ACLAPSFSFNYY224 pKa = 9.55IVPTNLLEE232 pKa = 4.29EE233 pKa = 4.62DD234 pKa = 4.22EE235 pKa = 4.62EE236 pKa = 4.73VLTFDD241 pKa = 5.53LDD243 pKa = 3.8PVPILSQFPEE253 pKa = 4.06LTEE256 pKa = 4.11LRR258 pKa = 11.84FHH260 pKa = 6.74ATSAMNGNQVVYY272 pKa = 10.07LNPLGSDD279 pKa = 2.84KK280 pKa = 11.3GLLKK284 pKa = 10.71EE285 pKa = 4.72PIRR288 pKa = 11.84TLVTTTTAIQGDD300 pKa = 3.89VSVPQSQSTTKK311 pKa = 10.19PPEE314 pKa = 3.92YY315 pKa = 9.59PPTVGSGKK323 pKa = 10.22KK324 pKa = 9.51VIEE327 pKa = 4.24YY328 pKa = 7.49EE329 pKa = 4.25TDD331 pKa = 3.16SSSCGLFCPNNNNPLPINTVTLVLDD356 pKa = 4.33GGAILIAAKK365 pKa = 8.98ITDD368 pKa = 3.52NTPRR372 pKa = 11.84SGVVSLGVDD381 pKa = 3.69GALTQVRR388 pKa = 11.84HH389 pKa = 5.81NSSTSQVITQVAPNGEE405 pKa = 4.38AIITFHH411 pKa = 6.53WKK413 pKa = 10.47GDD415 pKa = 3.69TEE417 pKa = 4.18QDD419 pKa = 4.26GVCMTPGMAKK429 pKa = 9.37QCQNIDD435 pKa = 3.7PSTGTPIYY443 pKa = 8.93TLRR446 pKa = 11.84DD447 pKa = 3.37AKK449 pKa = 10.11TDD451 pKa = 3.69QVLLWVRR458 pKa = 11.84LNYY461 pKa = 10.68NGFFSTTPTNVLTRR475 pKa = 11.84ISLANKK481 pKa = 8.49VLKK484 pKa = 10.54FEE486 pKa = 4.07YY487 pKa = 10.18RR488 pKa = 11.84LAITEE493 pKa = 4.21RR494 pKa = 11.84LPIPEE499 pKa = 4.27GSEE502 pKa = 3.95LNRR505 pKa = 11.84IAAEE509 pKa = 3.8QTSFVLADD517 pKa = 4.45LKK519 pKa = 11.19QEE521 pKa = 4.09MEE523 pKa = 4.5SKK525 pKa = 10.02FRR527 pKa = 11.84NYY529 pKa = 9.13FAAFLSRR536 pKa = 11.84HH537 pKa = 5.76PMSAHH542 pKa = 5.47DD543 pKa = 4.85HH544 pKa = 6.17DD545 pKa = 4.28WKK547 pKa = 11.06DD548 pKa = 3.41FEE550 pKa = 5.5DD551 pKa = 3.75RR552 pKa = 11.84VRR554 pKa = 11.84PLDD557 pKa = 3.92TPLLDD562 pKa = 4.45PKK564 pKa = 10.17LFCATTPFSAPDD576 pKa = 3.33QASTSLGRR584 pKa = 11.84QLSDD588 pKa = 2.99IGDD591 pKa = 3.58EE592 pKa = 4.82LEE594 pKa = 4.29YY595 pKa = 11.1SNCGHH600 pKa = 7.34RR601 pKa = 11.84DD602 pKa = 3.19CKK604 pKa = 10.15YY605 pKa = 11.09AKK607 pKa = 9.86SCLKK611 pKa = 10.92AEE613 pKa = 4.22TDD615 pKa = 3.27
MM1 pKa = 7.57SDD3 pKa = 4.1KK4 pKa = 10.8DD5 pKa = 3.42AAMPPTASGEE15 pKa = 4.04ITEE18 pKa = 4.66GMGVPPEE25 pKa = 4.13MPVDD29 pKa = 3.85AGIDD33 pKa = 3.46RR34 pKa = 11.84PGGITAEE41 pKa = 4.24VQQVHH46 pKa = 5.63TSRR49 pKa = 11.84NDD51 pKa = 3.34SIEE54 pKa = 3.88AFLRR58 pKa = 11.84SVPILQKK65 pKa = 10.13IIKK68 pKa = 9.18WSVNDD73 pKa = 3.67LPGTILYY80 pKa = 7.77QVPVHH85 pKa = 6.31PSYY88 pKa = 11.46VNYY91 pKa = 8.4MVKK94 pKa = 9.67WFKK97 pKa = 10.83KK98 pKa = 9.74AFKK101 pKa = 9.25YY102 pKa = 9.99WNGPIIFMIDD112 pKa = 3.04LAGTGFHH119 pKa = 6.53GGKK122 pKa = 10.26LIIFHH127 pKa = 6.4TPPGKK132 pKa = 10.03RR133 pKa = 11.84FKK135 pKa = 10.0PSKK138 pKa = 8.35MTMAQWFTFSHH149 pKa = 6.4GLVDD153 pKa = 5.84AKK155 pKa = 11.22SQGNNLACAPDD166 pKa = 3.86VIGRR170 pKa = 11.84TLHH173 pKa = 7.23DD174 pKa = 4.26NGPFDD179 pKa = 4.78EE180 pKa = 4.87NNEE183 pKa = 3.99DD184 pKa = 4.15TYY186 pKa = 11.73GGWFGIGVVMEE197 pKa = 5.1LKK199 pKa = 10.16TSGTGDD205 pKa = 3.36QEE207 pKa = 4.17IQFSLEE213 pKa = 3.57ACLAPSFSFNYY224 pKa = 9.55IVPTNLLEE232 pKa = 4.29EE233 pKa = 4.62DD234 pKa = 4.22EE235 pKa = 4.62EE236 pKa = 4.73VLTFDD241 pKa = 5.53LDD243 pKa = 3.8PVPILSQFPEE253 pKa = 4.06LTEE256 pKa = 4.11LRR258 pKa = 11.84FHH260 pKa = 6.74ATSAMNGNQVVYY272 pKa = 10.07LNPLGSDD279 pKa = 2.84KK280 pKa = 11.3GLLKK284 pKa = 10.71EE285 pKa = 4.72PIRR288 pKa = 11.84TLVTTTTAIQGDD300 pKa = 3.89VSVPQSQSTTKK311 pKa = 10.19PPEE314 pKa = 3.92YY315 pKa = 9.59PPTVGSGKK323 pKa = 10.22KK324 pKa = 9.51VIEE327 pKa = 4.24YY328 pKa = 7.49EE329 pKa = 4.25TDD331 pKa = 3.16SSSCGLFCPNNNNPLPINTVTLVLDD356 pKa = 4.33GGAILIAAKK365 pKa = 8.98ITDD368 pKa = 3.52NTPRR372 pKa = 11.84SGVVSLGVDD381 pKa = 3.69GALTQVRR388 pKa = 11.84HH389 pKa = 5.81NSSTSQVITQVAPNGEE405 pKa = 4.38AIITFHH411 pKa = 6.53WKK413 pKa = 10.47GDD415 pKa = 3.69TEE417 pKa = 4.18QDD419 pKa = 4.26GVCMTPGMAKK429 pKa = 9.37QCQNIDD435 pKa = 3.7PSTGTPIYY443 pKa = 8.93TLRR446 pKa = 11.84DD447 pKa = 3.37AKK449 pKa = 10.11TDD451 pKa = 3.69QVLLWVRR458 pKa = 11.84LNYY461 pKa = 10.68NGFFSTTPTNVLTRR475 pKa = 11.84ISLANKK481 pKa = 8.49VLKK484 pKa = 10.54FEE486 pKa = 4.07YY487 pKa = 10.18RR488 pKa = 11.84LAITEE493 pKa = 4.21RR494 pKa = 11.84LPIPEE499 pKa = 4.27GSEE502 pKa = 3.95LNRR505 pKa = 11.84IAAEE509 pKa = 3.8QTSFVLADD517 pKa = 4.45LKK519 pKa = 11.19QEE521 pKa = 4.09MEE523 pKa = 4.5SKK525 pKa = 10.02FRR527 pKa = 11.84NYY529 pKa = 9.13FAAFLSRR536 pKa = 11.84HH537 pKa = 5.76PMSAHH542 pKa = 5.47DD543 pKa = 4.85HH544 pKa = 6.17DD545 pKa = 4.28WKK547 pKa = 11.06DD548 pKa = 3.41FEE550 pKa = 5.5DD551 pKa = 3.75RR552 pKa = 11.84VRR554 pKa = 11.84PLDD557 pKa = 3.92TPLLDD562 pKa = 4.45PKK564 pKa = 10.17LFCATTPFSAPDD576 pKa = 3.33QASTSLGRR584 pKa = 11.84QLSDD588 pKa = 2.99IGDD591 pKa = 3.58EE592 pKa = 4.82LEE594 pKa = 4.29YY595 pKa = 11.1SNCGHH600 pKa = 7.34RR601 pKa = 11.84DD602 pKa = 3.19CKK604 pKa = 10.15YY605 pKa = 11.09AKK607 pKa = 9.86SCLKK611 pKa = 10.92AEE613 pKa = 4.22TDD615 pKa = 3.27
Molecular weight: 67.63 kDa
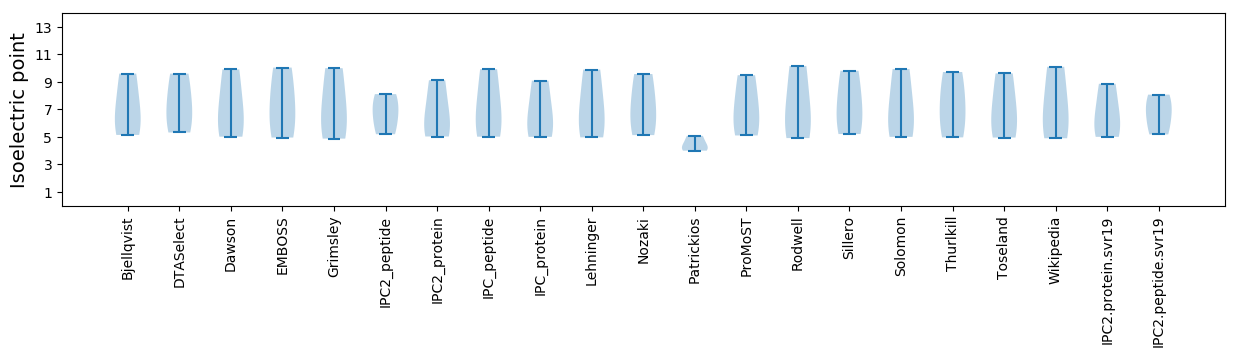
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KH81|A0A1L3KH81_9VIRU Uncharacterized protein OS=Beihai picorna-like virus 118 OX=1922547 PE=4 SV=1
MM1 pKa = 7.31AAAAAAAAGVKK12 pKa = 10.39AGGDD16 pKa = 3.75VVSTTTKK23 pKa = 10.7GLFDD27 pKa = 3.9FFQNRR32 pKa = 11.84EE33 pKa = 3.92NIDD36 pKa = 3.39YY37 pKa = 10.88KK38 pKa = 11.04KK39 pKa = 11.02FEE41 pKa = 4.58LATNQNNQLYY51 pKa = 8.09NTFLYY56 pKa = 9.57HH57 pKa = 7.05QNFFLPEE64 pKa = 4.07LEE66 pKa = 4.64LKK68 pKa = 10.44RR69 pKa = 11.84KK70 pKa = 8.53QVQSQYY76 pKa = 11.67DD77 pKa = 3.72VAALNQIEE85 pKa = 4.34SRR87 pKa = 11.84RR88 pKa = 11.84TAQQLSAYY96 pKa = 9.49NRR98 pKa = 11.84EE99 pKa = 3.93TALQLGQQNFSNALKK114 pKa = 10.63LGNNQFEE121 pKa = 4.42QSRR124 pKa = 11.84KK125 pKa = 9.94SMQLATKK132 pKa = 10.3LNLEE136 pKa = 4.6ADD138 pKa = 3.42VSAYY142 pKa = 7.79QRR144 pKa = 11.84KK145 pKa = 8.43LATQTNVLNRR155 pKa = 11.84DD156 pKa = 3.74GLPSSLAILGAGAPRR171 pKa = 11.84EE172 pKa = 3.88RR173 pKa = 11.84IYY175 pKa = 11.19AGNRR179 pKa = 11.84TFVSAYY185 pKa = 9.44QGNGPAYY192 pKa = 10.43SSTPSQMMLGNIKK205 pKa = 10.49LEE207 pKa = 4.38PKK209 pKa = 10.38
MM1 pKa = 7.31AAAAAAAAGVKK12 pKa = 10.39AGGDD16 pKa = 3.75VVSTTTKK23 pKa = 10.7GLFDD27 pKa = 3.9FFQNRR32 pKa = 11.84EE33 pKa = 3.92NIDD36 pKa = 3.39YY37 pKa = 10.88KK38 pKa = 11.04KK39 pKa = 11.02FEE41 pKa = 4.58LATNQNNQLYY51 pKa = 8.09NTFLYY56 pKa = 9.57HH57 pKa = 7.05QNFFLPEE64 pKa = 4.07LEE66 pKa = 4.64LKK68 pKa = 10.44RR69 pKa = 11.84KK70 pKa = 8.53QVQSQYY76 pKa = 11.67DD77 pKa = 3.72VAALNQIEE85 pKa = 4.34SRR87 pKa = 11.84RR88 pKa = 11.84TAQQLSAYY96 pKa = 9.49NRR98 pKa = 11.84EE99 pKa = 3.93TALQLGQQNFSNALKK114 pKa = 10.63LGNNQFEE121 pKa = 4.42QSRR124 pKa = 11.84KK125 pKa = 9.94SMQLATKK132 pKa = 10.3LNLEE136 pKa = 4.6ADD138 pKa = 3.42VSAYY142 pKa = 7.79QRR144 pKa = 11.84KK145 pKa = 8.43LATQTNVLNRR155 pKa = 11.84DD156 pKa = 3.74GLPSSLAILGAGAPRR171 pKa = 11.84EE172 pKa = 3.88RR173 pKa = 11.84IYY175 pKa = 11.19AGNRR179 pKa = 11.84TFVSAYY185 pKa = 9.44QGNGPAYY192 pKa = 10.43SSTPSQMMLGNIKK205 pKa = 10.49LEE207 pKa = 4.38PKK209 pKa = 10.38
Molecular weight: 23.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2837 |
209 |
2013 |
945.7 |
106.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.768 ± 1.57 | 1.692 ± 0.479 |
6.591 ± 1.063 | 5.675 ± 0.303 |
4.935 ± 0.113 | 5.746 ± 0.737 |
2.82 ± 0.769 | 5.816 ± 0.968 |
6.768 ± 0.866 | 9.059 ± 0.571 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.939 ± 0.151 | 4.406 ± 1.358 |
5.746 ± 0.988 | 3.913 ± 1.582 |
4.582 ± 0.531 | 7.191 ± 0.175 |
6.838 ± 0.679 | 5.146 ± 0.604 |
1.128 ± 0.314 | 3.243 ± 0.432 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
