
Arabidopsis halleri partitivirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
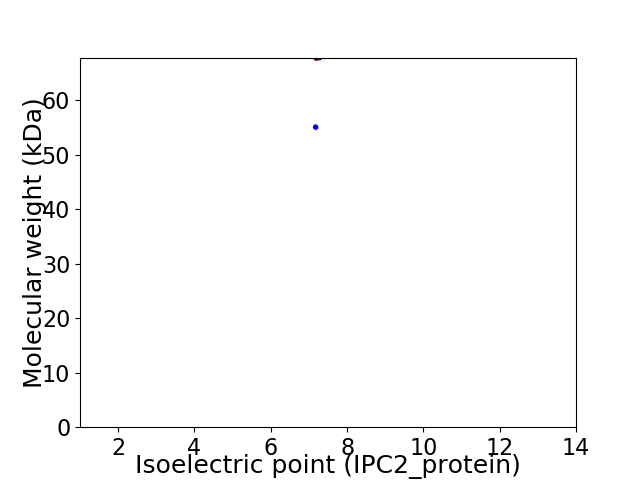
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B4ZBZ5|A0A1B4ZBZ5_9VIRU RNA-dependent RNA polymerase OS=Arabidopsis halleri partitivirus 1 OX=1849335 GN=RdRP PE=4 SV=1
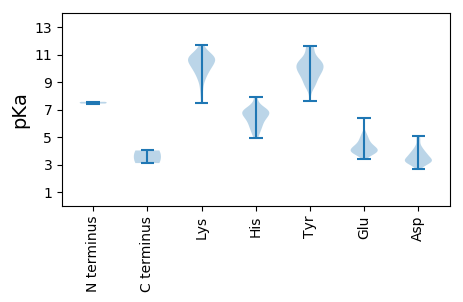
MM1 pKa = 7.43SMKK4 pKa = 10.24RR5 pKa = 11.84KK6 pKa = 9.39SRR8 pKa = 11.84PSSSTKK14 pKa = 10.38DD15 pKa = 3.41LEE17 pKa = 5.09VEE19 pKa = 3.96DD20 pKa = 5.1TYY22 pKa = 11.65LKK24 pKa = 10.72QSGLDD29 pKa = 3.42SMNKK33 pKa = 9.91LEE35 pKa = 4.82PVEE38 pKa = 3.93QSKK41 pKa = 11.44DD42 pKa = 3.37EE43 pKa = 4.21EE44 pKa = 4.54TTKK47 pKa = 10.88VSMLPTASTAIIAPRR62 pKa = 11.84KK63 pKa = 8.23LTAEE67 pKa = 4.39DD68 pKa = 4.62FSSKK72 pKa = 10.24RR73 pKa = 11.84KK74 pKa = 9.19PDD76 pKa = 3.21QTSAVSPFFGFLRR89 pKa = 11.84THH91 pKa = 7.03ILHH94 pKa = 6.42PTQGRR99 pKa = 11.84LSHH102 pKa = 6.45YY103 pKa = 10.22YY104 pKa = 9.41PSCHH108 pKa = 5.63MMDD111 pKa = 4.8YY112 pKa = 10.11ILHH115 pKa = 6.77SINSTLCDD123 pKa = 2.82NYY125 pKa = 10.2YY126 pKa = 10.63FKK128 pKa = 11.15RR129 pKa = 11.84EE130 pKa = 3.91TPNYY134 pKa = 8.87HH135 pKa = 6.8PYY137 pKa = 10.43ILRR140 pKa = 11.84LYY142 pKa = 10.19FGVLFWVQCLRR153 pKa = 11.84AGNDD157 pKa = 3.18VQVINDD163 pKa = 3.34LHH165 pKa = 7.93YY166 pKa = 11.44DD167 pKa = 4.01FLQRR171 pKa = 11.84FLDD174 pKa = 4.13CNPLEE179 pKa = 4.41SLAIPGPLLGLFKK192 pKa = 9.67TLCSSQPEE200 pKa = 4.17FPHH203 pKa = 5.86YY204 pKa = 10.63GKK206 pKa = 10.64VYY208 pKa = 9.91PRR210 pKa = 11.84IPASPGPRR218 pKa = 11.84RR219 pKa = 11.84RR220 pKa = 11.84DD221 pKa = 3.25MFSKK225 pKa = 10.47NVPSAQFLPNVPGIFALIHH244 pKa = 5.98HH245 pKa = 6.77LHH247 pKa = 6.2GLSEE251 pKa = 4.65GEE253 pKa = 3.83HH254 pKa = 6.64PIYY257 pKa = 10.23PKK259 pKa = 10.2RR260 pKa = 11.84KK261 pKa = 7.47RR262 pKa = 11.84HH263 pKa = 5.46IPVTEE268 pKa = 3.76EE269 pKa = 3.43ASNFGFKK276 pKa = 10.56AFAAFPNRR284 pKa = 11.84IQRR287 pKa = 11.84DD288 pKa = 3.17RR289 pKa = 11.84WMVSSPGLQYY299 pKa = 10.71PCEE302 pKa = 4.66ADD304 pKa = 2.91MKK306 pKa = 9.69MNEE309 pKa = 3.76AFAEE313 pKa = 4.02RR314 pKa = 11.84FYY316 pKa = 11.57DD317 pKa = 3.77FDD319 pKa = 4.21FPAFNADD326 pKa = 4.32DD327 pKa = 4.07NLSTITNFLHH337 pKa = 5.97MRR339 pKa = 11.84KK340 pKa = 9.92SMAWFIRR347 pKa = 11.84AKK349 pKa = 10.3EE350 pKa = 4.26VACSAARR357 pKa = 11.84FFSDD361 pKa = 3.16SGTLADD367 pKa = 4.81CSPHH371 pKa = 7.26GLVSNQIIVAITPPPEE387 pKa = 3.72EE388 pKa = 4.13TFADD392 pKa = 3.89PRR394 pKa = 11.84FSADD398 pKa = 3.13PRR400 pKa = 11.84ALYY403 pKa = 9.71PFSFKK408 pKa = 10.86LKK410 pKa = 9.08STAHH414 pKa = 6.25NLPPLAEE421 pKa = 3.95AAAAFSQTHH430 pKa = 5.05IRR432 pKa = 11.84IFPEE436 pKa = 3.82YY437 pKa = 9.33PLAGNFGQKK446 pKa = 8.36TDD448 pKa = 3.4EE449 pKa = 5.36SGPFWDD455 pKa = 4.14IRR457 pKa = 11.84PIGSSPTDD465 pKa = 3.26DD466 pKa = 3.27TSYY469 pKa = 10.27LTIPPMVKK477 pKa = 9.91AALIEE482 pKa = 4.54KK483 pKa = 10.19GSSRR487 pKa = 4.02
MM1 pKa = 7.43SMKK4 pKa = 10.24RR5 pKa = 11.84KK6 pKa = 9.39SRR8 pKa = 11.84PSSSTKK14 pKa = 10.38DD15 pKa = 3.41LEE17 pKa = 5.09VEE19 pKa = 3.96DD20 pKa = 5.1TYY22 pKa = 11.65LKK24 pKa = 10.72QSGLDD29 pKa = 3.42SMNKK33 pKa = 9.91LEE35 pKa = 4.82PVEE38 pKa = 3.93QSKK41 pKa = 11.44DD42 pKa = 3.37EE43 pKa = 4.21EE44 pKa = 4.54TTKK47 pKa = 10.88VSMLPTASTAIIAPRR62 pKa = 11.84KK63 pKa = 8.23LTAEE67 pKa = 4.39DD68 pKa = 4.62FSSKK72 pKa = 10.24RR73 pKa = 11.84KK74 pKa = 9.19PDD76 pKa = 3.21QTSAVSPFFGFLRR89 pKa = 11.84THH91 pKa = 7.03ILHH94 pKa = 6.42PTQGRR99 pKa = 11.84LSHH102 pKa = 6.45YY103 pKa = 10.22YY104 pKa = 9.41PSCHH108 pKa = 5.63MMDD111 pKa = 4.8YY112 pKa = 10.11ILHH115 pKa = 6.77SINSTLCDD123 pKa = 2.82NYY125 pKa = 10.2YY126 pKa = 10.63FKK128 pKa = 11.15RR129 pKa = 11.84EE130 pKa = 3.91TPNYY134 pKa = 8.87HH135 pKa = 6.8PYY137 pKa = 10.43ILRR140 pKa = 11.84LYY142 pKa = 10.19FGVLFWVQCLRR153 pKa = 11.84AGNDD157 pKa = 3.18VQVINDD163 pKa = 3.34LHH165 pKa = 7.93YY166 pKa = 11.44DD167 pKa = 4.01FLQRR171 pKa = 11.84FLDD174 pKa = 4.13CNPLEE179 pKa = 4.41SLAIPGPLLGLFKK192 pKa = 9.67TLCSSQPEE200 pKa = 4.17FPHH203 pKa = 5.86YY204 pKa = 10.63GKK206 pKa = 10.64VYY208 pKa = 9.91PRR210 pKa = 11.84IPASPGPRR218 pKa = 11.84RR219 pKa = 11.84RR220 pKa = 11.84DD221 pKa = 3.25MFSKK225 pKa = 10.47NVPSAQFLPNVPGIFALIHH244 pKa = 5.98HH245 pKa = 6.77LHH247 pKa = 6.2GLSEE251 pKa = 4.65GEE253 pKa = 3.83HH254 pKa = 6.64PIYY257 pKa = 10.23PKK259 pKa = 10.2RR260 pKa = 11.84KK261 pKa = 7.47RR262 pKa = 11.84HH263 pKa = 5.46IPVTEE268 pKa = 3.76EE269 pKa = 3.43ASNFGFKK276 pKa = 10.56AFAAFPNRR284 pKa = 11.84IQRR287 pKa = 11.84DD288 pKa = 3.17RR289 pKa = 11.84WMVSSPGLQYY299 pKa = 10.71PCEE302 pKa = 4.66ADD304 pKa = 2.91MKK306 pKa = 9.69MNEE309 pKa = 3.76AFAEE313 pKa = 4.02RR314 pKa = 11.84FYY316 pKa = 11.57DD317 pKa = 3.77FDD319 pKa = 4.21FPAFNADD326 pKa = 4.32DD327 pKa = 4.07NLSTITNFLHH337 pKa = 5.97MRR339 pKa = 11.84KK340 pKa = 9.92SMAWFIRR347 pKa = 11.84AKK349 pKa = 10.3EE350 pKa = 4.26VACSAARR357 pKa = 11.84FFSDD361 pKa = 3.16SGTLADD367 pKa = 4.81CSPHH371 pKa = 7.26GLVSNQIIVAITPPPEE387 pKa = 3.72EE388 pKa = 4.13TFADD392 pKa = 3.89PRR394 pKa = 11.84FSADD398 pKa = 3.13PRR400 pKa = 11.84ALYY403 pKa = 9.71PFSFKK408 pKa = 10.86LKK410 pKa = 9.08STAHH414 pKa = 6.25NLPPLAEE421 pKa = 3.95AAAAFSQTHH430 pKa = 5.05IRR432 pKa = 11.84IFPEE436 pKa = 3.82YY437 pKa = 9.33PLAGNFGQKK446 pKa = 8.36TDD448 pKa = 3.4EE449 pKa = 5.36SGPFWDD455 pKa = 4.14IRR457 pKa = 11.84PIGSSPTDD465 pKa = 3.26DD466 pKa = 3.27TSYY469 pKa = 10.27LTIPPMVKK477 pKa = 9.91AALIEE482 pKa = 4.54KK483 pKa = 10.19GSSRR487 pKa = 4.02
Molecular weight: 55.1 kDa
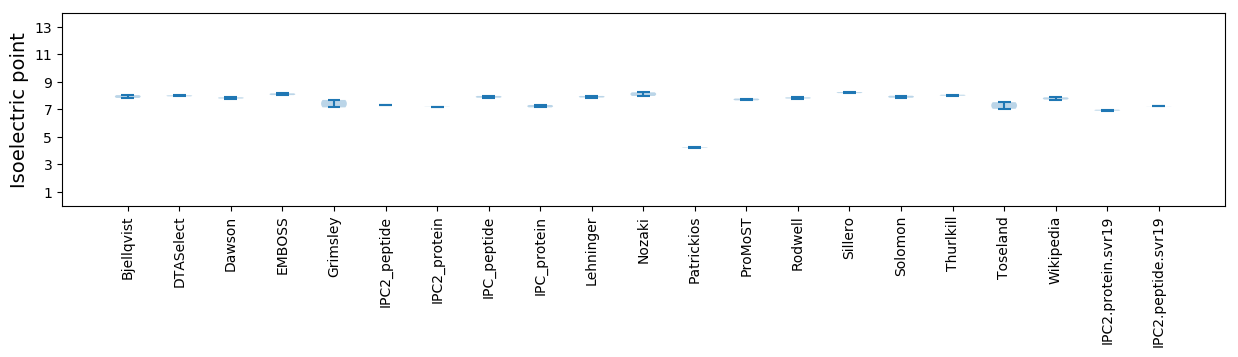
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B4ZBZ5|A0A1B4ZBZ5_9VIRU RNA-dependent RNA polymerase OS=Arabidopsis halleri partitivirus 1 OX=1849335 GN=RdRP PE=4 SV=1
MM1 pKa = 7.56KK2 pKa = 9.43NTVVLEE8 pKa = 4.09PLPSLARR15 pKa = 11.84PIYY18 pKa = 10.49GDD20 pKa = 3.55TDD22 pKa = 3.77PGRR25 pKa = 11.84NPAYY29 pKa = 10.31QSTVDD34 pKa = 3.16HH35 pKa = 6.79ALRR38 pKa = 11.84RR39 pKa = 11.84LLTAEE44 pKa = 3.96EE45 pKa = 4.14FNIVVNGYY53 pKa = 9.44RR54 pKa = 11.84RR55 pKa = 11.84SPWNEE60 pKa = 3.72DD61 pKa = 2.95ALTADD66 pKa = 3.57IEE68 pKa = 4.44KK69 pKa = 10.85LNSDD73 pKa = 3.74YY74 pKa = 11.62HH75 pKa = 6.85HH76 pKa = 6.71VNKK79 pKa = 10.51DD80 pKa = 2.94EE81 pKa = 4.72HH82 pKa = 6.65YY83 pKa = 9.72YY84 pKa = 10.74KK85 pKa = 10.63AIEE88 pKa = 4.11HH89 pKa = 5.6TKK91 pKa = 10.89KK92 pKa = 10.78LFTPKK97 pKa = 10.17EE98 pKa = 3.93KK99 pKa = 10.52LRR101 pKa = 11.84PVHH104 pKa = 6.84FNDD107 pKa = 3.23LRR109 pKa = 11.84HH110 pKa = 5.85YY111 pKa = 8.77PWQLSTSIGAPFATSEE127 pKa = 3.82KK128 pKa = 8.85WKK130 pKa = 10.72DD131 pKa = 3.83YY132 pKa = 11.16INQKK136 pKa = 10.46YY137 pKa = 10.05DD138 pKa = 3.17GKK140 pKa = 11.0LKK142 pKa = 10.99SRR144 pKa = 11.84DD145 pKa = 3.55FKK147 pKa = 11.7DD148 pKa = 3.52LFKK151 pKa = 9.88EE152 pKa = 4.04THH154 pKa = 5.7GVSLEE159 pKa = 3.84PYY161 pKa = 8.96MIDD164 pKa = 3.32RR165 pKa = 11.84RR166 pKa = 11.84LSKK169 pKa = 10.8RR170 pKa = 11.84NFYY173 pKa = 11.49NEE175 pKa = 3.38MFYY178 pKa = 10.66INRR181 pKa = 11.84INIHH185 pKa = 6.42HH186 pKa = 6.94IKK188 pKa = 10.66DD189 pKa = 3.26GWTTNPAGHH198 pKa = 7.47DD199 pKa = 3.22LRR201 pKa = 11.84YY202 pKa = 8.96WHH204 pKa = 6.54TAHH207 pKa = 7.24ARR209 pKa = 11.84QHH211 pKa = 5.72LVEE214 pKa = 6.38AGDD217 pKa = 3.69EE218 pKa = 4.35DD219 pKa = 4.26KK220 pKa = 11.53VRR222 pKa = 11.84LVFGAPSTLLMAEE235 pKa = 5.16LMFIWPIQTSLLARR249 pKa = 11.84GSSSPMLWGYY259 pKa = 7.61EE260 pKa = 4.06TTTGGWSRR268 pKa = 11.84LYY270 pKa = 10.23NWAYY274 pKa = 10.43SALPRR279 pKa = 11.84FGAVATLDD287 pKa = 3.16WSRR290 pKa = 11.84FDD292 pKa = 3.12KK293 pKa = 10.81DD294 pKa = 3.13ARR296 pKa = 11.84HH297 pKa = 5.2TVITDD302 pKa = 2.81IHH304 pKa = 6.94DD305 pKa = 4.97LIMRR309 pKa = 11.84PMFDD313 pKa = 3.84FNSGYY318 pKa = 10.28HH319 pKa = 4.96PTIINPRR326 pKa = 11.84SNPDD330 pKa = 3.29PQRR333 pKa = 11.84LEE335 pKa = 4.85NLWNWMKK342 pKa = 10.84NAILTTPLLLPDD354 pKa = 3.76GTRR357 pKa = 11.84LQFQHH362 pKa = 6.68SGIYY366 pKa = 9.69SGYY369 pKa = 9.65FQTQILDD376 pKa = 3.45SMYY379 pKa = 8.75NCVMIFTVLSRR390 pKa = 11.84MGFDD394 pKa = 3.97LNSVAIKK401 pKa = 10.63VQGDD405 pKa = 3.5DD406 pKa = 3.71SLILLSHH413 pKa = 7.04SYY415 pKa = 9.96TFLQHH420 pKa = 6.93SFLTTFAHH428 pKa = 6.42HH429 pKa = 6.22AAVYY433 pKa = 9.76FGSTLNVKK441 pKa = 10.18KK442 pKa = 10.91SEE444 pKa = 4.25LLPSLEE450 pKa = 4.12DD451 pKa = 3.72AEE453 pKa = 4.33VLRR456 pKa = 11.84YY457 pKa = 9.56RR458 pKa = 11.84NHH460 pKa = 6.97GMMPYY465 pKa = 9.77RR466 pKa = 11.84EE467 pKa = 4.22EE468 pKa = 4.12LQLLAMLRR476 pKa = 11.84HH477 pKa = 6.2PEE479 pKa = 3.89RR480 pKa = 11.84TASLSALMARR490 pKa = 11.84SIGIAYY496 pKa = 8.7ANCGNYY502 pKa = 8.65TRR504 pKa = 11.84VHH506 pKa = 6.83HH507 pKa = 6.31ICEE510 pKa = 5.19DD511 pKa = 2.66IHH513 pKa = 7.94NYY515 pKa = 10.23LKK517 pKa = 11.13GIGVKK522 pKa = 9.77PDD524 pKa = 3.22AFGLPGGLRR533 pKa = 11.84FRR535 pKa = 11.84KK536 pKa = 9.87NYY538 pKa = 9.55LPSYY542 pKa = 9.24EE543 pKa = 4.96EE544 pKa = 3.73IDD546 pKa = 3.32ISHH549 pKa = 7.06FPTWLEE555 pKa = 3.95TVEE558 pKa = 4.88RR559 pKa = 11.84LLDD562 pKa = 3.69PSRR565 pKa = 11.84PLLTNKK571 pKa = 9.32HH572 pKa = 5.29WPTTHH577 pKa = 6.79FFGIPGEE584 pKa = 4.37SS585 pKa = 3.11
MM1 pKa = 7.56KK2 pKa = 9.43NTVVLEE8 pKa = 4.09PLPSLARR15 pKa = 11.84PIYY18 pKa = 10.49GDD20 pKa = 3.55TDD22 pKa = 3.77PGRR25 pKa = 11.84NPAYY29 pKa = 10.31QSTVDD34 pKa = 3.16HH35 pKa = 6.79ALRR38 pKa = 11.84RR39 pKa = 11.84LLTAEE44 pKa = 3.96EE45 pKa = 4.14FNIVVNGYY53 pKa = 9.44RR54 pKa = 11.84RR55 pKa = 11.84SPWNEE60 pKa = 3.72DD61 pKa = 2.95ALTADD66 pKa = 3.57IEE68 pKa = 4.44KK69 pKa = 10.85LNSDD73 pKa = 3.74YY74 pKa = 11.62HH75 pKa = 6.85HH76 pKa = 6.71VNKK79 pKa = 10.51DD80 pKa = 2.94EE81 pKa = 4.72HH82 pKa = 6.65YY83 pKa = 9.72YY84 pKa = 10.74KK85 pKa = 10.63AIEE88 pKa = 4.11HH89 pKa = 5.6TKK91 pKa = 10.89KK92 pKa = 10.78LFTPKK97 pKa = 10.17EE98 pKa = 3.93KK99 pKa = 10.52LRR101 pKa = 11.84PVHH104 pKa = 6.84FNDD107 pKa = 3.23LRR109 pKa = 11.84HH110 pKa = 5.85YY111 pKa = 8.77PWQLSTSIGAPFATSEE127 pKa = 3.82KK128 pKa = 8.85WKK130 pKa = 10.72DD131 pKa = 3.83YY132 pKa = 11.16INQKK136 pKa = 10.46YY137 pKa = 10.05DD138 pKa = 3.17GKK140 pKa = 11.0LKK142 pKa = 10.99SRR144 pKa = 11.84DD145 pKa = 3.55FKK147 pKa = 11.7DD148 pKa = 3.52LFKK151 pKa = 9.88EE152 pKa = 4.04THH154 pKa = 5.7GVSLEE159 pKa = 3.84PYY161 pKa = 8.96MIDD164 pKa = 3.32RR165 pKa = 11.84RR166 pKa = 11.84LSKK169 pKa = 10.8RR170 pKa = 11.84NFYY173 pKa = 11.49NEE175 pKa = 3.38MFYY178 pKa = 10.66INRR181 pKa = 11.84INIHH185 pKa = 6.42HH186 pKa = 6.94IKK188 pKa = 10.66DD189 pKa = 3.26GWTTNPAGHH198 pKa = 7.47DD199 pKa = 3.22LRR201 pKa = 11.84YY202 pKa = 8.96WHH204 pKa = 6.54TAHH207 pKa = 7.24ARR209 pKa = 11.84QHH211 pKa = 5.72LVEE214 pKa = 6.38AGDD217 pKa = 3.69EE218 pKa = 4.35DD219 pKa = 4.26KK220 pKa = 11.53VRR222 pKa = 11.84LVFGAPSTLLMAEE235 pKa = 5.16LMFIWPIQTSLLARR249 pKa = 11.84GSSSPMLWGYY259 pKa = 7.61EE260 pKa = 4.06TTTGGWSRR268 pKa = 11.84LYY270 pKa = 10.23NWAYY274 pKa = 10.43SALPRR279 pKa = 11.84FGAVATLDD287 pKa = 3.16WSRR290 pKa = 11.84FDD292 pKa = 3.12KK293 pKa = 10.81DD294 pKa = 3.13ARR296 pKa = 11.84HH297 pKa = 5.2TVITDD302 pKa = 2.81IHH304 pKa = 6.94DD305 pKa = 4.97LIMRR309 pKa = 11.84PMFDD313 pKa = 3.84FNSGYY318 pKa = 10.28HH319 pKa = 4.96PTIINPRR326 pKa = 11.84SNPDD330 pKa = 3.29PQRR333 pKa = 11.84LEE335 pKa = 4.85NLWNWMKK342 pKa = 10.84NAILTTPLLLPDD354 pKa = 3.76GTRR357 pKa = 11.84LQFQHH362 pKa = 6.68SGIYY366 pKa = 9.69SGYY369 pKa = 9.65FQTQILDD376 pKa = 3.45SMYY379 pKa = 8.75NCVMIFTVLSRR390 pKa = 11.84MGFDD394 pKa = 3.97LNSVAIKK401 pKa = 10.63VQGDD405 pKa = 3.5DD406 pKa = 3.71SLILLSHH413 pKa = 7.04SYY415 pKa = 9.96TFLQHH420 pKa = 6.93SFLTTFAHH428 pKa = 6.42HH429 pKa = 6.22AAVYY433 pKa = 9.76FGSTLNVKK441 pKa = 10.18KK442 pKa = 10.91SEE444 pKa = 4.25LLPSLEE450 pKa = 4.12DD451 pKa = 3.72AEE453 pKa = 4.33VLRR456 pKa = 11.84YY457 pKa = 9.56RR458 pKa = 11.84NHH460 pKa = 6.97GMMPYY465 pKa = 9.77RR466 pKa = 11.84EE467 pKa = 4.22EE468 pKa = 4.12LQLLAMLRR476 pKa = 11.84HH477 pKa = 6.2PEE479 pKa = 3.89RR480 pKa = 11.84TASLSALMARR490 pKa = 11.84SIGIAYY496 pKa = 8.7ANCGNYY502 pKa = 8.65TRR504 pKa = 11.84VHH506 pKa = 6.83HH507 pKa = 6.31ICEE510 pKa = 5.19DD511 pKa = 2.66IHH513 pKa = 7.94NYY515 pKa = 10.23LKK517 pKa = 11.13GIGVKK522 pKa = 9.77PDD524 pKa = 3.22AFGLPGGLRR533 pKa = 11.84FRR535 pKa = 11.84KK536 pKa = 9.87NYY538 pKa = 9.55LPSYY542 pKa = 9.24EE543 pKa = 4.96EE544 pKa = 3.73IDD546 pKa = 3.32ISHH549 pKa = 7.06FPTWLEE555 pKa = 3.95TVEE558 pKa = 4.88RR559 pKa = 11.84LLDD562 pKa = 3.69PSRR565 pKa = 11.84PLLTNKK571 pKa = 9.32HH572 pKa = 5.29WPTTHH577 pKa = 6.79FFGIPGEE584 pKa = 4.37SS585 pKa = 3.11
Molecular weight: 67.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1072 |
487 |
585 |
536.0 |
61.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.716 ± 0.645 | 1.026 ± 0.366 |
5.69 ± 0.087 | 4.944 ± 0.009 |
5.784 ± 0.833 | 4.944 ± 0.375 |
4.384 ± 0.531 | 5.317 ± 0.109 |
4.851 ± 0.168 | 9.701 ± 0.883 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.705 ± 0.021 | 4.478 ± 0.464 |
7.369 ± 1.111 | 2.519 ± 0.211 |
6.063 ± 0.186 | 7.836 ± 0.834 |
5.877 ± 0.441 | 3.731 ± 0.143 |
1.679 ± 0.509 | 4.384 ± 0.409 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
