
Gordonia phage GAL1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Galunavirus; Gordonia virus GAL1
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
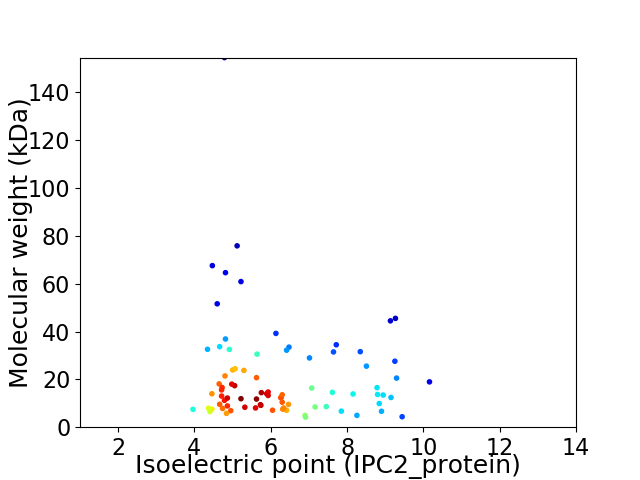
Virtual 2D-PAGE plot for 82 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
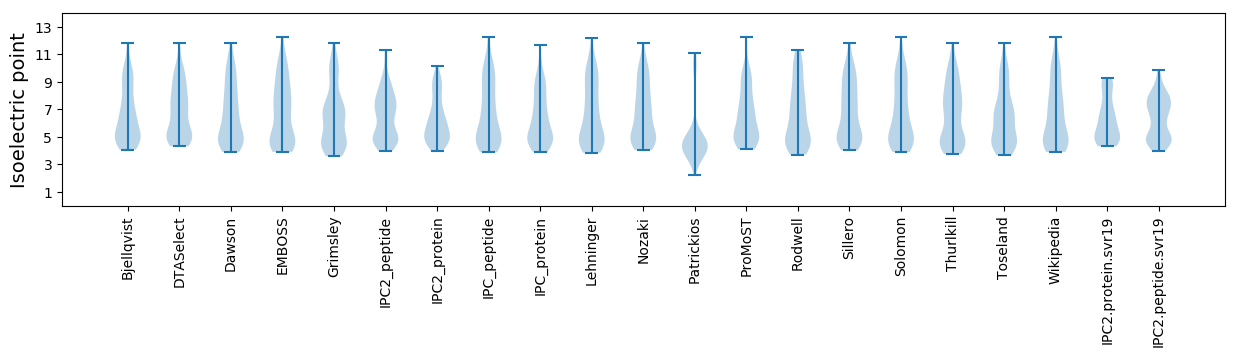
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A159B6G5|A0A159B6G5_9CAUD Uncharacterized protein OS=Gordonia phage GAL1 OX=1647469 GN=GAL1_79 PE=4 SV=1
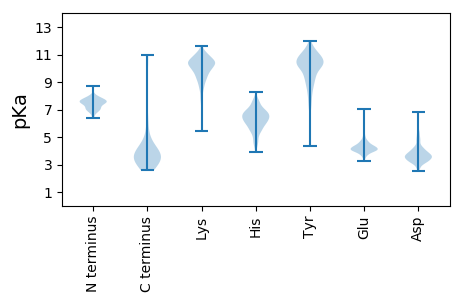
MM1 pKa = 8.5RR2 pKa = 11.84MMCFINPDD10 pKa = 2.83GSKK13 pKa = 10.41RR14 pKa = 11.84YY15 pKa = 9.86VNPLQVSVIEE25 pKa = 4.21KK26 pKa = 9.55TDD28 pKa = 3.59DD29 pKa = 3.4GGVRR33 pKa = 11.84MIYY36 pKa = 9.92PGGYY40 pKa = 9.93VLFPDD45 pKa = 4.85GDD47 pKa = 3.76IDD49 pKa = 4.1QLASQWFHH57 pKa = 6.14SVNAVIDD64 pKa = 4.15PSEE67 pKa = 4.52LEE69 pKa = 4.06APP71 pKa = 4.01
MM1 pKa = 8.5RR2 pKa = 11.84MMCFINPDD10 pKa = 2.83GSKK13 pKa = 10.41RR14 pKa = 11.84YY15 pKa = 9.86VNPLQVSVIEE25 pKa = 4.21KK26 pKa = 9.55TDD28 pKa = 3.59DD29 pKa = 3.4GGVRR33 pKa = 11.84MIYY36 pKa = 9.92PGGYY40 pKa = 9.93VLFPDD45 pKa = 4.85GDD47 pKa = 3.76IDD49 pKa = 4.1QLASQWFHH57 pKa = 6.14SVNAVIDD64 pKa = 4.15PSEE67 pKa = 4.52LEE69 pKa = 4.06APP71 pKa = 4.01
Molecular weight: 7.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A159B6E9|A0A159B6E9_9CAUD Uncharacterized protein OS=Gordonia phage GAL1 OX=1647469 GN=GAL1_54 PE=4 SV=1
MM1 pKa = 7.1STGFHH6 pKa = 6.41IEE8 pKa = 4.12YY9 pKa = 10.23VPSRR13 pKa = 11.84NMDD16 pKa = 3.0RR17 pKa = 11.84RR18 pKa = 11.84SDD20 pKa = 3.46RR21 pKa = 11.84NTIDD25 pKa = 4.13DD26 pKa = 3.8KK27 pKa = 11.25KK28 pKa = 10.41QAVGSSSVIPGSNLLNRR45 pKa = 11.84NRR47 pKa = 11.84NVSEE51 pKa = 4.11MLADD55 pKa = 4.26LPKK58 pKa = 10.32PPAWHH63 pKa = 6.56EE64 pKa = 3.91LGICRR69 pKa = 11.84QFDD72 pKa = 3.64PEE74 pKa = 3.94IFYY77 pKa = 10.11PSHH80 pKa = 6.42YY81 pKa = 10.67GPTQTADD88 pKa = 3.26AKK90 pKa = 10.4KK91 pKa = 9.88VCAGCDD97 pKa = 3.35VRR99 pKa = 11.84AVCLQWALDD108 pKa = 3.7NNDD111 pKa = 3.0QHH113 pKa = 6.89GVLGGTTPRR122 pKa = 11.84EE123 pKa = 4.39SEE125 pKa = 3.99SHH127 pKa = 5.56GQGGCGMTCQCGRR140 pKa = 11.84PAHH143 pKa = 6.1ARR145 pKa = 11.84GRR147 pKa = 11.84CARR150 pKa = 11.84CYY152 pKa = 9.25AQLRR156 pKa = 11.84ARR158 pKa = 11.84QTAYY162 pKa = 10.53GRR164 pKa = 11.84WEE166 pKa = 3.92PSYY169 pKa = 11.38TDD171 pKa = 3.34AQPVRR176 pKa = 11.84EE177 pKa = 4.38HH178 pKa = 6.77LLALRR183 pKa = 11.84AAGIGNRR190 pKa = 11.84KK191 pKa = 8.6VRR193 pKa = 11.84EE194 pKa = 4.13LTVIAPSTIQAILYY208 pKa = 7.29GRR210 pKa = 11.84PARR213 pKa = 11.84GNPPARR219 pKa = 11.84KK220 pKa = 9.34VLRR223 pKa = 11.84NTAARR228 pKa = 11.84VLAIPVPVSKK238 pKa = 10.82VPCLRR243 pKa = 11.84GGRR246 pKa = 11.84VVPALGSVRR255 pKa = 11.84RR256 pKa = 11.84LQALAANGYY265 pKa = 7.09SQRR268 pKa = 11.84EE269 pKa = 3.97LWSRR273 pKa = 11.84LGWPSPQNATQMFAGRR289 pKa = 11.84VEE291 pKa = 4.51NITVVRR297 pKa = 11.84AQQVTVLFSQLQMVPGTDD315 pKa = 2.72RR316 pKa = 11.84NARR319 pKa = 11.84ARR321 pKa = 11.84AKK323 pKa = 10.69AKK325 pKa = 10.14GWLPPLAWDD334 pKa = 4.33EE335 pKa = 4.93DD336 pKa = 4.8RR337 pKa = 11.84IDD339 pKa = 4.31DD340 pKa = 3.8PTYY343 pKa = 10.43TPEE346 pKa = 4.95LVDD349 pKa = 3.82KK350 pKa = 10.43PRR352 pKa = 11.84TAEE355 pKa = 4.04DD356 pKa = 3.56VFSDD360 pKa = 4.01FEE362 pKa = 4.44YY363 pKa = 10.69LLSMGVGVEE372 pKa = 4.02DD373 pKa = 3.8ASRR376 pKa = 11.84RR377 pKa = 11.84VGLLPASVKK386 pKa = 9.98RR387 pKa = 11.84RR388 pKa = 11.84YY389 pKa = 8.35EE390 pKa = 3.7RR391 pKa = 11.84HH392 pKa = 5.39GRR394 pKa = 11.84RR395 pKa = 11.84CPAALTAVARR405 pKa = 11.84QQRR408 pKa = 11.84KK409 pKa = 5.42TASS412 pKa = 3.14
MM1 pKa = 7.1STGFHH6 pKa = 6.41IEE8 pKa = 4.12YY9 pKa = 10.23VPSRR13 pKa = 11.84NMDD16 pKa = 3.0RR17 pKa = 11.84RR18 pKa = 11.84SDD20 pKa = 3.46RR21 pKa = 11.84NTIDD25 pKa = 4.13DD26 pKa = 3.8KK27 pKa = 11.25KK28 pKa = 10.41QAVGSSSVIPGSNLLNRR45 pKa = 11.84NRR47 pKa = 11.84NVSEE51 pKa = 4.11MLADD55 pKa = 4.26LPKK58 pKa = 10.32PPAWHH63 pKa = 6.56EE64 pKa = 3.91LGICRR69 pKa = 11.84QFDD72 pKa = 3.64PEE74 pKa = 3.94IFYY77 pKa = 10.11PSHH80 pKa = 6.42YY81 pKa = 10.67GPTQTADD88 pKa = 3.26AKK90 pKa = 10.4KK91 pKa = 9.88VCAGCDD97 pKa = 3.35VRR99 pKa = 11.84AVCLQWALDD108 pKa = 3.7NNDD111 pKa = 3.0QHH113 pKa = 6.89GVLGGTTPRR122 pKa = 11.84EE123 pKa = 4.39SEE125 pKa = 3.99SHH127 pKa = 5.56GQGGCGMTCQCGRR140 pKa = 11.84PAHH143 pKa = 6.1ARR145 pKa = 11.84GRR147 pKa = 11.84CARR150 pKa = 11.84CYY152 pKa = 9.25AQLRR156 pKa = 11.84ARR158 pKa = 11.84QTAYY162 pKa = 10.53GRR164 pKa = 11.84WEE166 pKa = 3.92PSYY169 pKa = 11.38TDD171 pKa = 3.34AQPVRR176 pKa = 11.84EE177 pKa = 4.38HH178 pKa = 6.77LLALRR183 pKa = 11.84AAGIGNRR190 pKa = 11.84KK191 pKa = 8.6VRR193 pKa = 11.84EE194 pKa = 4.13LTVIAPSTIQAILYY208 pKa = 7.29GRR210 pKa = 11.84PARR213 pKa = 11.84GNPPARR219 pKa = 11.84KK220 pKa = 9.34VLRR223 pKa = 11.84NTAARR228 pKa = 11.84VLAIPVPVSKK238 pKa = 10.82VPCLRR243 pKa = 11.84GGRR246 pKa = 11.84VVPALGSVRR255 pKa = 11.84RR256 pKa = 11.84LQALAANGYY265 pKa = 7.09SQRR268 pKa = 11.84EE269 pKa = 3.97LWSRR273 pKa = 11.84LGWPSPQNATQMFAGRR289 pKa = 11.84VEE291 pKa = 4.51NITVVRR297 pKa = 11.84AQQVTVLFSQLQMVPGTDD315 pKa = 2.72RR316 pKa = 11.84NARR319 pKa = 11.84ARR321 pKa = 11.84AKK323 pKa = 10.69AKK325 pKa = 10.14GWLPPLAWDD334 pKa = 4.33EE335 pKa = 4.93DD336 pKa = 4.8RR337 pKa = 11.84IDD339 pKa = 4.31DD340 pKa = 3.8PTYY343 pKa = 10.43TPEE346 pKa = 4.95LVDD349 pKa = 3.82KK350 pKa = 10.43PRR352 pKa = 11.84TAEE355 pKa = 4.04DD356 pKa = 3.56VFSDD360 pKa = 4.01FEE362 pKa = 4.44YY363 pKa = 10.69LLSMGVGVEE372 pKa = 4.02DD373 pKa = 3.8ASRR376 pKa = 11.84RR377 pKa = 11.84VGLLPASVKK386 pKa = 9.98RR387 pKa = 11.84RR388 pKa = 11.84YY389 pKa = 8.35EE390 pKa = 3.7RR391 pKa = 11.84HH392 pKa = 5.39GRR394 pKa = 11.84RR395 pKa = 11.84CPAALTAVARR405 pKa = 11.84QQRR408 pKa = 11.84KK409 pKa = 5.42TASS412 pKa = 3.14
Molecular weight: 45.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
15857 |
38 |
1465 |
193.4 |
21.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.187 ± 0.437 | 1.173 ± 0.205 |
6.685 ± 0.287 | 5.651 ± 0.274 |
2.882 ± 0.173 | 8.186 ± 0.352 |
2.245 ± 0.249 | 4.257 ± 0.168 |
3.241 ± 0.232 | 7.858 ± 0.298 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.207 ± 0.143 | 2.964 ± 0.182 |
5.966 ± 0.178 | 3.847 ± 0.333 |
7.669 ± 0.437 | 5.531 ± 0.299 |
6.842 ± 0.312 | 7.315 ± 0.273 |
1.98 ± 0.137 | 2.314 ± 0.141 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
